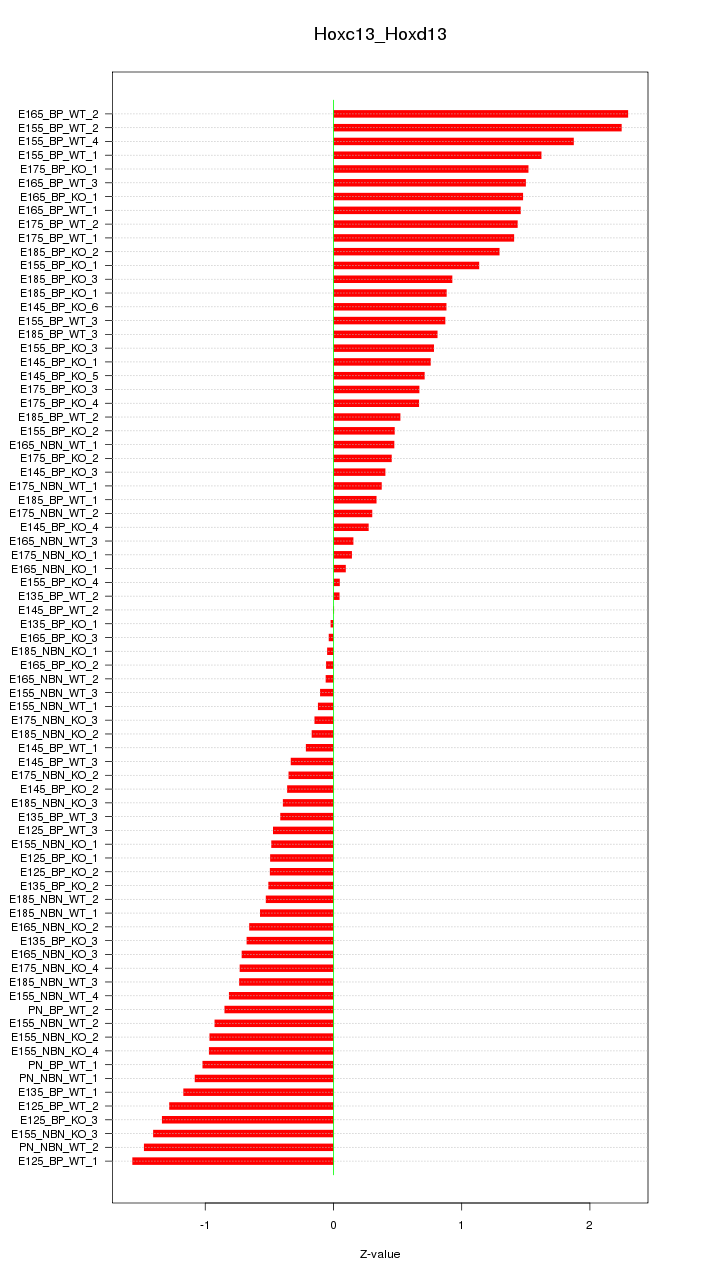
Motif ID: Hoxc13_Hoxd13
Z-value: 0.906
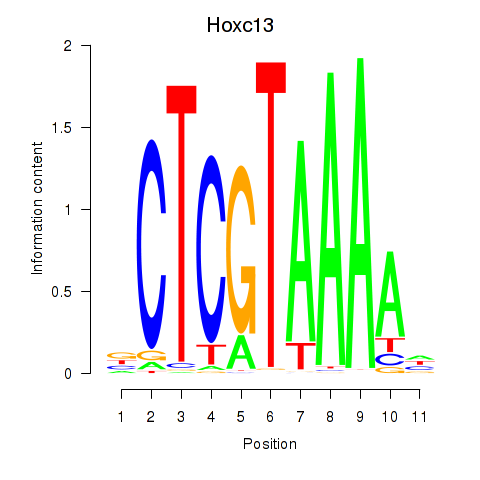
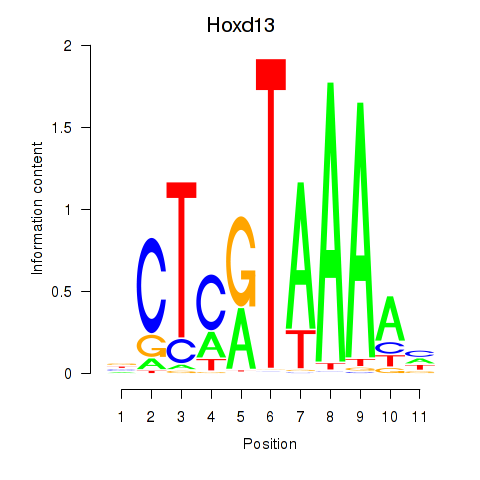
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 6.9 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 1.8 | 11.0 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 1.3 | 7.9 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 1.2 | 4.7 | GO:2000110 | protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 1.1 | 6.3 | GO:0009405 | pathogenesis(GO:0009405) |
| 1.0 | 5.2 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 1.0 | 4.0 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) negative regulation of t-circle formation(GO:1904430) |
| 1.0 | 26.7 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.8 | 7.5 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.7 | 2.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 | 4.0 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.7 | 7.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.5 | 2.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.4 | 1.3 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.4 | 7.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.3 | 4.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.3 | 1.3 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.2 | 2.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 4.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.2 | 1.1 | GO:0071472 | cellular response to salt stress(GO:0071472) |
| 0.2 | 2.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 1.9 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 0.3 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) |
| 0.1 | 0.5 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 19.0 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.1 | 6.4 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 0.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 3.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.9 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 1.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 4.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 1.6 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 2.9 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 0.8 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.2 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.2 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.3 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.0 | 3.6 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 1.1 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.2 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 5.1 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.2 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 1.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 26.3 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 2.6 | 7.9 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 1.0 | 4.0 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.7 | 2.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 4.7 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.4 | 5.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 3.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 6.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 4.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 14.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 2.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.3 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.3 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 3.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 4.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 1.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 4.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 31.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 2.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0035102 | PRC1 complex(GO:0035102) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 26.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 2.6 | 7.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.8 | 7.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.9 | 4.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.9 | 6.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.8 | 11.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.6 | 5.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.5 | 6.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.5 | 22.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.4 | 4.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.4 | 1.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 7.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 4.0 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.3 | 2.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.3 | 0.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.3 | 1.3 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.2 | 3.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 5.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 0.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 1.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 0.6 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.4 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 2.0 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.1 | 3.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 5.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 2.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.3 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.1 | 3.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.3 | GO:0043426 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) MRF binding(GO:0043426) |
| 0.1 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 4.3 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 5.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.4 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 6.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.8 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.4 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 26.3 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.0 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 5.2 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.1 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 6.0 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 7.5 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 4.4 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.1 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 5.6 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.0 | 0.4 | PID_ARF6_PATHWAY | Arf6 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.7 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 22.7 | REACTOME_CHONDROITIN_SULFATE_DERMATAN_SULFATE_METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.3 | 5.2 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.2 | 4.0 | REACTOME_INTEGRATION_OF_PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 0.6 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 7.2 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 8.2 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 5.0 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 7.5 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.4 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.5 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.2 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.8 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 2.8 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.1 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |