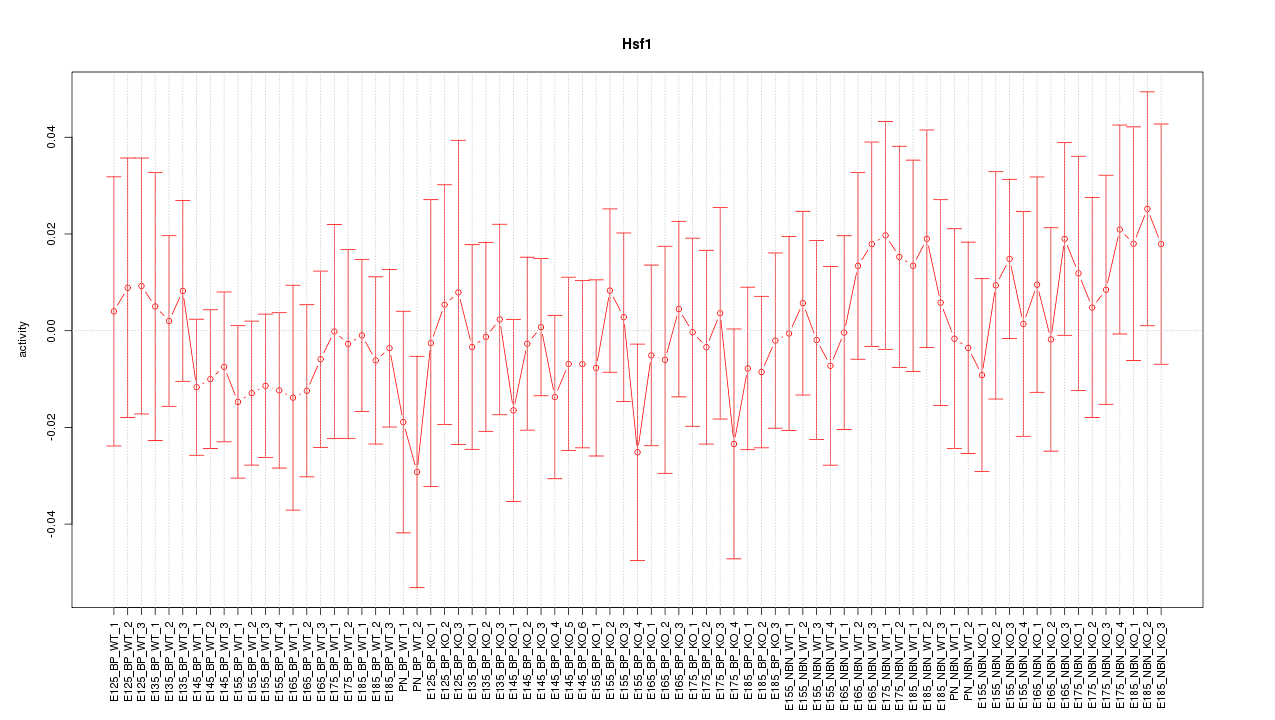
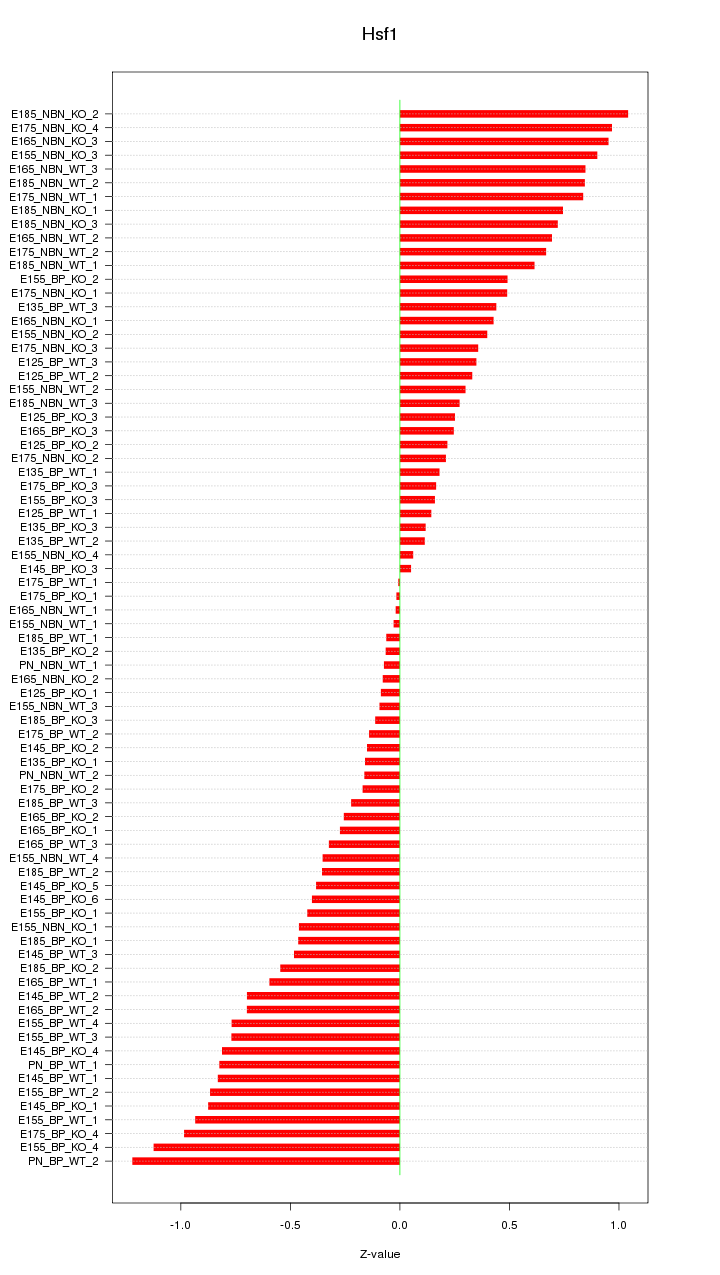
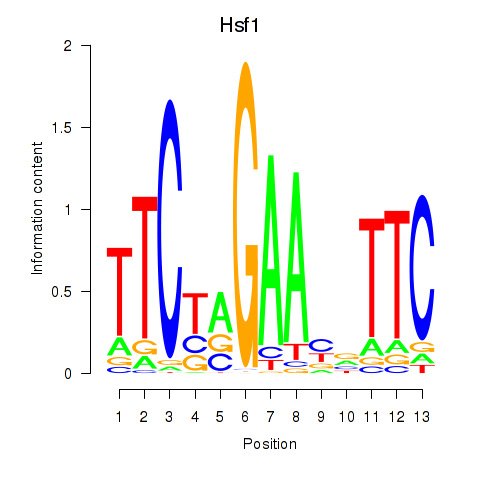
Motif ID: Hsf1
Z-value: 0.546
Transcription factors associated with Hsf1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hsf1 | ENSMUSG00000022556.9 | Hsf1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf1 | mm10_v2_chr15_+_76477404_76477461 | -0.25 | 2.8e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.5 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 1.0 | 2.9 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 1.0 | 3.9 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.9 | 2.6 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.7 | 5.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.6 | 3.6 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.6 | 3.5 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.5 | 5.8 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.5 | 5.0 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.5 | 1.5 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.5 | 2.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.4 | 1.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.3 | 3.7 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.3 | 2.2 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.3 | 1.9 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.2 | 3.4 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.2 | 1.0 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.2 | 0.7 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.2 | 1.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 0.7 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 | 2.9 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 1.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.3 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 | 1.1 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 3.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.1 | 0.6 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 3.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.2 | GO:0019062 | virion attachment to host cell(GO:0019062) transforming growth factor beta activation(GO:0036363) adhesion of symbiont to host cell(GO:0044650) complement-dependent cytotoxicity(GO:0097278) |
| 0.1 | 0.4 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 1.2 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 2.3 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.7 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.0 | 0.5 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.7 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.1 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) embryonic brain development(GO:1990403) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.5 | 5.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.4 | 3.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 1.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 7.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 3.5 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 4.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.6 | GO:0043209 | myelin sheath(GO:0043209) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | GO:0032564 | dATP binding(GO:0032564) |
| 0.6 | 3.7 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 3.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.5 | 2.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.4 | 3.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.4 | 1.6 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.3 | 3.9 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 1.0 | GO:0070140 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.3 | 2.6 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 5.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.5 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 1.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 1.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 5.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.4 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 2.9 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 4.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.9 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 2.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 7.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.3 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.1 | 7.5 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.6 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.5 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 2.0 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.9 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.0 | 3.9 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.2 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.5 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.7 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.5 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 5.0 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 5.8 | REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 3.5 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.6 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.9 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.3 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.5 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 3.8 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |