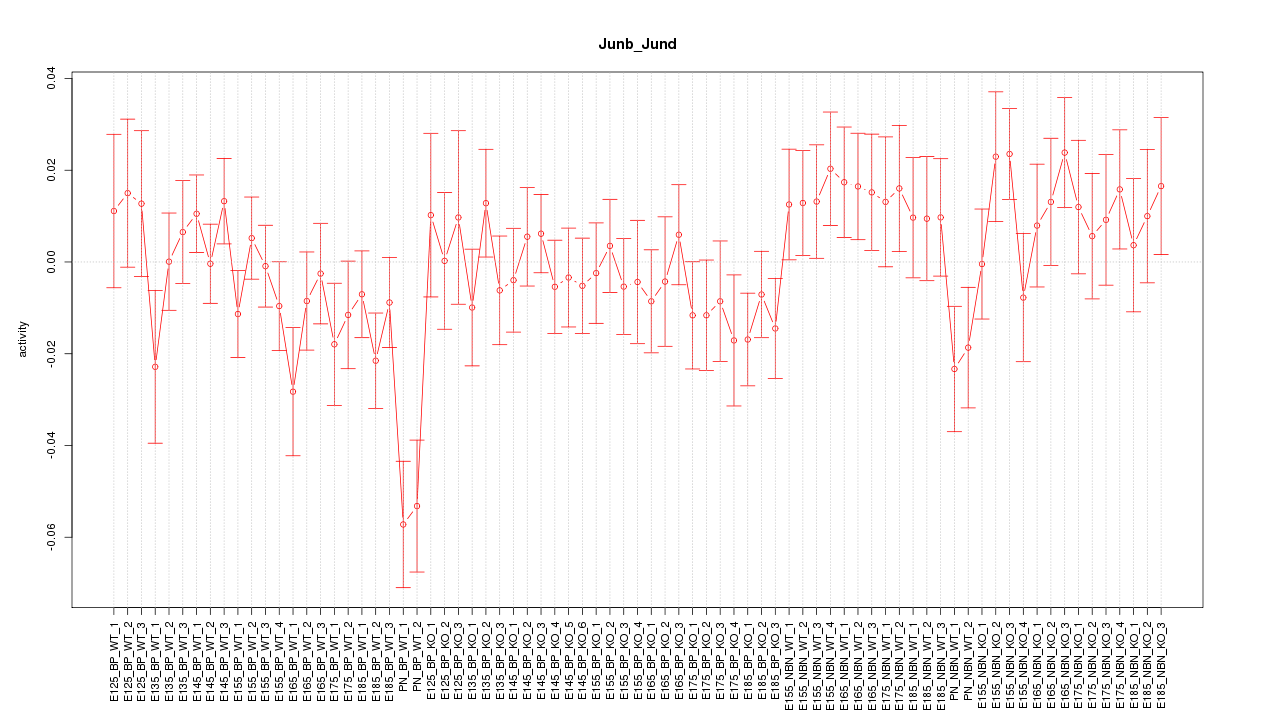
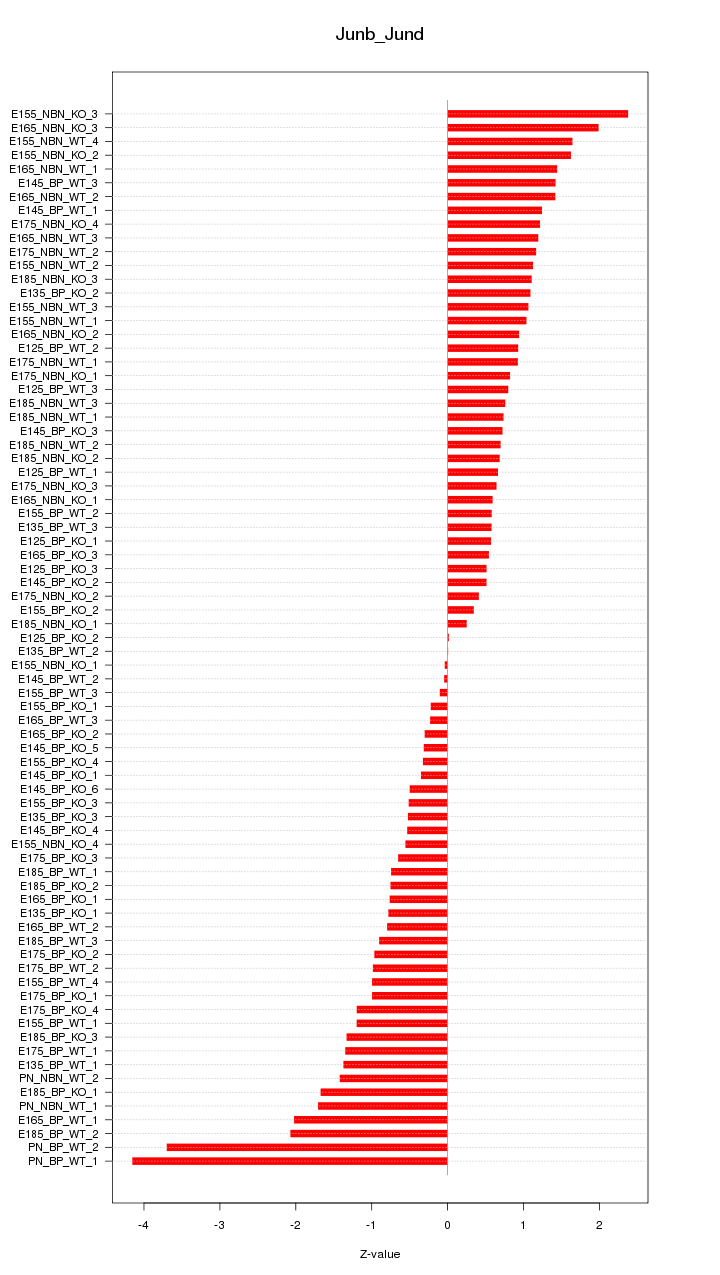
Motif ID: Junb_Jund
Z-value: 1.185

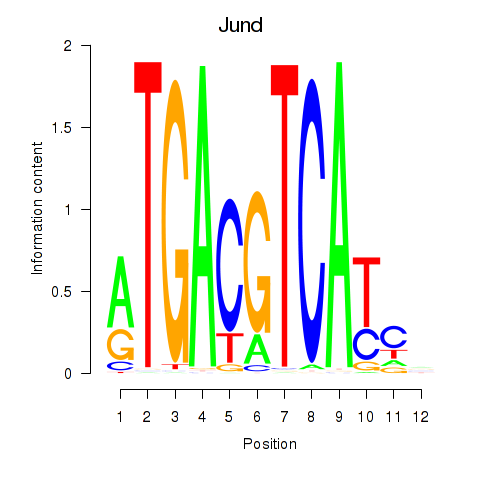
Transcription factors associated with Junb_Jund:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Junb | ENSMUSG00000052837.5 | Junb |
| Jund | ENSMUSG00000071076.5 | Jund |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Junb | mm10_v2_chr8_-_84978709_84978748 | 0.35 | 1.5e-03 | Click! |
| Jund | mm10_v2_chr8_+_70697739_70697739 | 0.01 | 9.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.5 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 5.0 | 14.9 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) transforming growth factor-beta secretion(GO:0038044) |
| 3.5 | 10.4 | GO:0030070 | insulin processing(GO:0030070) |
| 2.9 | 8.8 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 2.8 | 8.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 2.5 | 9.9 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 2.2 | 6.7 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 2.2 | 13.4 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 2.1 | 12.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 2.1 | 24.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 1.9 | 9.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 1.9 | 9.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 1.5 | 10.3 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 1.4 | 13.0 | GO:0060373 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.4 | 4.3 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 1.4 | 5.6 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 1.3 | 5.3 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 1.3 | 1.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 1.3 | 7.6 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 1.2 | 3.7 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 1.2 | 4.7 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 1.1 | 1.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 1.1 | 10.0 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.1 | 3.3 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.0 | 9.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.0 | 6.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.0 | 4.9 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 1.0 | 2.9 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 1.0 | 4.8 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.9 | 6.5 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.9 | 0.9 | GO:0060067 | cervix development(GO:0060067) |
| 0.9 | 7.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.9 | 11.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.8 | 4.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.8 | 1.6 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.8 | 1.6 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.8 | 4.0 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.8 | 3.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.8 | 2.3 | GO:0071336 | regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) actin filament branching(GO:0090135) |
| 0.8 | 4.5 | GO:0060702 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.8 | 10.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.7 | 5.9 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.7 | 2.2 | GO:0043379 | memory T cell differentiation(GO:0043379) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.7 | 3.6 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.7 | 5.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.7 | 4.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.7 | 2.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.7 | 18.0 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.7 | 9.7 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.7 | 5.4 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.7 | 2.0 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 0.7 | 2.0 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.6 | 3.9 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.6 | 2.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.6 | 5.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.6 | 1.9 | GO:0071649 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of somatostatin secretion(GO:0090274) |
| 0.6 | 0.6 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.6 | 1.8 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.6 | 1.2 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.6 | 1.8 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.6 | 4.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.6 | 2.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.5 | 1.6 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.5 | 11.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.5 | 2.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.5 | 1.6 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.5 | 2.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.5 | 1.5 | GO:0019389 | urate transport(GO:0015747) glucuronoside metabolic process(GO:0019389) negative regulation of intestinal absorption(GO:1904479) |
| 0.5 | 2.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.5 | 3.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.5 | 5.4 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.5 | 2.5 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.5 | 1.4 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.5 | 6.7 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.5 | 4.7 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.5 | 1.4 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.5 | 1.4 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.5 | 18.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.5 | 9.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.5 | 1.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.5 | 1.4 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.5 | 1.8 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.4 | 1.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.4 | 4.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 | 1.3 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.4 | 1.3 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.4 | 1.7 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.4 | 3.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.4 | 2.0 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.4 | 2.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 1.2 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.4 | 1.9 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.4 | 2.6 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.4 | 1.1 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.4 | 35.7 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.4 | 1.8 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.4 | 7.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 1.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 0.7 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.3 | 4.0 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.3 | 1.7 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.3 | 1.0 | GO:1903538 | meiotic sister chromatid cohesion, centromeric(GO:0051754) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.3 | 2.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.3 | 2.9 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 4.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.3 | 1.9 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.3 | 1.9 | GO:0014060 | regulation of epinephrine secretion(GO:0014060) |
| 0.3 | 1.6 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.3 | 1.6 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.3 | 0.9 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.3 | 0.9 | GO:0014028 | notochord formation(GO:0014028) axial mesoderm morphogenesis(GO:0048319) |
| 0.3 | 3.3 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.3 | 1.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 2.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.3 | 2.6 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.3 | 4.0 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.3 | 2.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.3 | 3.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.3 | 2.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.3 | 7.1 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.3 | 1.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 1.9 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) growth plate cartilage chondrocyte proliferation(GO:0003419) negative regulation of oocyte maturation(GO:1900194) |
| 0.3 | 1.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.3 | 5.8 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.3 | 6.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.3 | 0.5 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 1.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.2 | 2.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 20.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 7.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 0.7 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.2 | 1.7 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 1.7 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.2 | 1.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 3.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.2 | 0.9 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.2 | 0.9 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.2 | 0.7 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.2 | 1.4 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.2 | 1.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.2 | 1.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.2 | 1.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 1.7 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.2 | 4.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 0.4 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 | 2.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.2 | 2.8 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 2.0 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 3.5 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 1.2 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.2 | 0.8 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.2 | 2.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.2 | 1.3 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.2 | 0.6 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.2 | 0.5 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.2 | 0.7 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.2 | 0.5 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.2 | 0.4 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.2 | 1.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 1.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.2 | 1.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 2.3 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.2 | 0.7 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 0.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.2 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.2 | 0.5 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 0.8 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 0.5 | GO:2000192 | negative regulation of fatty acid transport(GO:2000192) |
| 0.2 | 0.8 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.2 | 0.8 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 1.6 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 0.5 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.2 | 4.8 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.2 | 0.8 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.2 | 1.4 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.2 | 0.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 0.6 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 1.9 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 1.0 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 5.7 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 1.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.2 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.1 | 5.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 0.4 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 2.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.7 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.5 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 | 4.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.5 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 0.8 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 1.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.4 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 2.0 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.7 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 9.0 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.1 | 0.6 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 1.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 1.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.8 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 1.6 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.1 | 3.5 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 3.7 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 0.3 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 2.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.9 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 9.5 | GO:0007613 | memory(GO:0007613) |
| 0.1 | 1.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.5 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.1 | 1.1 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.4 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.9 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 0.3 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.1 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 1.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.2 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.1 | 0.6 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.7 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 3.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 2.7 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 1.0 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 1.0 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.1 | 0.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.7 | GO:0021612 | facial nerve structural organization(GO:0021612) |
| 0.1 | 2.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.4 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.4 | GO:0018377 | N-terminal protein lipidation(GO:0006498) N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.1 | 0.7 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 2.6 | GO:0007215 | glutamate receptor signaling pathway(GO:0007215) |
| 0.1 | 0.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 1.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 10.0 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 0.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.2 | GO:0060192 | negative regulation of lipase activity(GO:0060192) |
| 0.1 | 1.3 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 2.9 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 0.6 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.1 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.1 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.1 | 1.1 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.1 | 0.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 1.8 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 2.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.6 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 2.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 3.0 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.9 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 2.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 3.8 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.1 | 3.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 1.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 1.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.8 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.7 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.6 | GO:0030575 | male sex determination(GO:0030238) nuclear body organization(GO:0030575) |
| 0.0 | 0.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.4 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 5.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 1.7 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.7 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 1.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 2.8 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.4 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.5 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.6 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.0 | 0.3 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.2 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) specification of loop of Henle identity(GO:0072086) |
| 0.0 | 2.0 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.7 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 3.6 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.9 | GO:0035150 | regulation of tube size(GO:0035150) regulation of blood vessel size(GO:0050880) |
| 0.0 | 0.2 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.3 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.1 | GO:0098885 | actin filament bundle distribution(GO:0070650) modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.0 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.7 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 2.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.4 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 11.6 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.4 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.3 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.5 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 3.1 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.0 | 2.0 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.5 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.0 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 14.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.6 | 1.6 | GO:0097447 | dendritic tree(GO:0097447) |
| 1.4 | 6.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.3 | 20.2 | GO:0044754 | autolysosome(GO:0044754) |
| 1.2 | 3.6 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 1.2 | 4.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.9 | 13.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.9 | 2.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.8 | 5.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.8 | 19.6 | GO:0031430 | M band(GO:0031430) |
| 0.7 | 4.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.6 | 4.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.6 | 3.8 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.6 | 1.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.6 | 1.8 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.6 | 2.3 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.6 | 2.3 | GO:0031673 | H zone(GO:0031673) |
| 0.5 | 7.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.4 | 9.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.4 | 1.7 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 3.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 1.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.4 | 5.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 1.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.3 | 5.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 12.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.3 | 2.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.3 | 4.6 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.3 | 6.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.3 | 1.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.3 | 0.9 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.3 | 2.2 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.3 | 5.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.3 | 2.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 11.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 1.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.3 | 0.5 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 3.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 4.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 11.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 1.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 0.9 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 15.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 7.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.2 | 8.9 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 3.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 2.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 8.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 8.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 1.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.2 | 12.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 2.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 1.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 2.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.7 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.2 | 1.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.2 | 3.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 8.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 8.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 3.6 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.2 | 1.8 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 0.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 1.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 0.6 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 1.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.9 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.9 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 2.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 43.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 12.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 8.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 6.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 18.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 2.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 2.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 2.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 3.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.1 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.8 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 1.1 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 4.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.6 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.1 | 1.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 8.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.7 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 2.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 2.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.8 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 2.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 0.4 | GO:0005655 | ribonuclease MRP complex(GO:0000172) nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 12.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 7.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 2.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 5.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.1 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 3.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.4 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.1 | 0.3 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 3.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 2.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.2 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 3.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 1.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 7.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 37.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 3.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.9 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 2.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.7 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 3.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 5.9 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.5 | GO:0019867 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.1 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.1 | GO:0044433 | cytoplasmic vesicle part(GO:0044433) |
| 0.0 | 0.1 | GO:0031011 | Ino80 complex(GO:0031011) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 14.9 | GO:0045340 | mercury ion binding(GO:0045340) |
| 2.4 | 19.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 2.2 | 6.7 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 1.9 | 13.0 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.7 | 17.0 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 1.6 | 4.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 1.6 | 11.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 1.4 | 11.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.4 | 4.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.2 | 3.7 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 1.2 | 9.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 1.1 | 4.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.1 | 3.3 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.0 | 15.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 1.0 | 10.6 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 1.0 | 2.9 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.9 | 14.0 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.9 | 7.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.9 | 3.5 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.9 | 2.6 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.8 | 9.5 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.8 | 4.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.8 | 15.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.7 | 5.9 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.7 | 2.9 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.7 | 7.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.7 | 4.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 2.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.6 | 10.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.6 | 2.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 4.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.6 | 1.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.6 | 7.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.6 | 8.4 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.6 | 3.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.6 | 1.7 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.6 | 18.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.5 | 3.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.5 | 2.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.5 | 1.4 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.5 | 2.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 1.9 | GO:0035473 | lipase binding(GO:0035473) |
| 0.5 | 1.4 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.5 | 3.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.5 | 1.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.5 | 2.3 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.4 | 3.6 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.4 | 1.3 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.4 | 2.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.4 | 1.2 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.4 | 2.7 | GO:0032564 | dATP binding(GO:0032564) |
| 0.4 | 1.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.4 | 1.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 7.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 5.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.3 | 2.0 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.3 | 2.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 6.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.3 | 1.0 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.3 | 1.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 1.6 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.3 | 3.7 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.3 | 1.5 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.3 | 3.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 1.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.3 | 6.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 1.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 1.9 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 2.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.3 | 4.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.3 | 0.8 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.3 | 8.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.3 | 2.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.3 | 1.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.2 | 4.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 0.9 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.2 | 0.9 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 15.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 2.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 5.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 3.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 2.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 3.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 2.2 | GO:0044213 | intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 4.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 1.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 0.9 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.2 | 5.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 0.6 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.2 | 2.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.7 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 1.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 4.3 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 1.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 2.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.2 | 4.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.2 | 8.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.2 | 11.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 1.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.3 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 11.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 4.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.6 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.4 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.1 | 16.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 8.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 4.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 11.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.4 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.4 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 1.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 2.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 2.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 2.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 4.9 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.1 | 4.9 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.3 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 1.8 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 1.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 33.9 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.6 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 27.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 1.1 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 3.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 2.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 3.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 5.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.5 | GO:0004043 | L-aminoadipate-semialdehyde dehydrogenase activity(GO:0004043) |
| 0.1 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 9.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 3.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 4.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 4.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 2.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 5.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 3.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.6 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 1.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 3.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.8 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.1 | 2.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.0 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 1.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 1.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 3.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 7.2 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.4 | GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen(GO:0016705) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 3.3 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 14.9 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.6 | 12.7 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.6 | 25.4 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.4 | 11.2 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 12.9 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 16.2 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.3 | 8.3 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 2.3 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 6.6 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 15.8 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.2 | 1.6 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.2 | 3.9 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 7.3 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.0 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.1 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.1 | 5.5 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.7 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 7.9 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.1 | 4.7 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.1 | 7.5 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 0.4 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 3.0 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.9 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.1 | 1.5 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 0.6 | PID_PI3K_PLC_TRK_PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 0.8 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.7 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.9 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.7 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.2 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.1 | 2.0 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 2.5 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.5 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.1 | 0.3 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.7 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.1 | 0.6 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 6.4 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.1 | 0.4 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.9 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.8 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.0 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.5 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.9 | PID_IL8_CXCR2_PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.8 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.7 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.6 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.8 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.1 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 14.8 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 1.0 | 2.0 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.9 | 18.2 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.9 | 30.9 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.7 | 4.9 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.6 | 13.4 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.5 | 20.6 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.5 | 8.6 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.5 | 7.3 | REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.5 | 10.0 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.5 | 7.5 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.4 | 2.9 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.3 | 5.1 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.3 | 8.3 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.3 | 1.6 | REACTOME_SIGNALLING_TO_P38_VIA_RIT_AND_RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.3 | 11.9 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 4.3 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 7.2 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 8.3 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 3.1 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 2.8 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 1.9 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 5.6 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 5.7 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 7.5 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 2.0 | REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 2.9 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 4.2 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 13.6 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 2.1 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.2 | 3.7 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 3.4 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 1.4 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 4.3 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 1.4 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 1.5 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.9 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 8.3 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.6 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.5 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 3.6 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.1 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 2.5 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 0.7 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.5 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.6 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.9 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.2 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.8 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 6.6 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 3.3 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 0.9 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 1.2 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.9 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.7 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.4 | REACTOME_ENERGY_DEPENDENT_REGULATION_OF_MTOR_BY_LKB1_AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.1 | 1.7 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.5 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.8 | REACTOME_REGULATION_OF_GLUCOKINASE_BY_GLUCOKINASE_REGULATORY_PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 0.6 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.9 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.6 | REACTOME_IL_3_5_AND_GM_CSF_SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.0 | 1.6 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.0 | 1.7 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.5 | REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 1.1 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.4 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 2.7 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.3 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 1.3 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.1 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.6 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.6 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME_PYRUVATE_METABOLISM_AND_CITRIC_ACID_TCA_CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.4 | REACTOME_AUTODEGRADATION_OF_CDH1_BY_CDH1_APC_C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.3 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.8 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.0 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.9 | REACTOME_HOST_INTERACTIONS_OF_HIV_FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.3 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME_NONSENSE_MEDIATED_DECAY_ENHANCED_BY_THE_EXON_JUNCTION_COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.4 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 2.3 | REACTOME_TRANSLATION | Genes involved in Translation |
| 0.0 | 0.2 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |