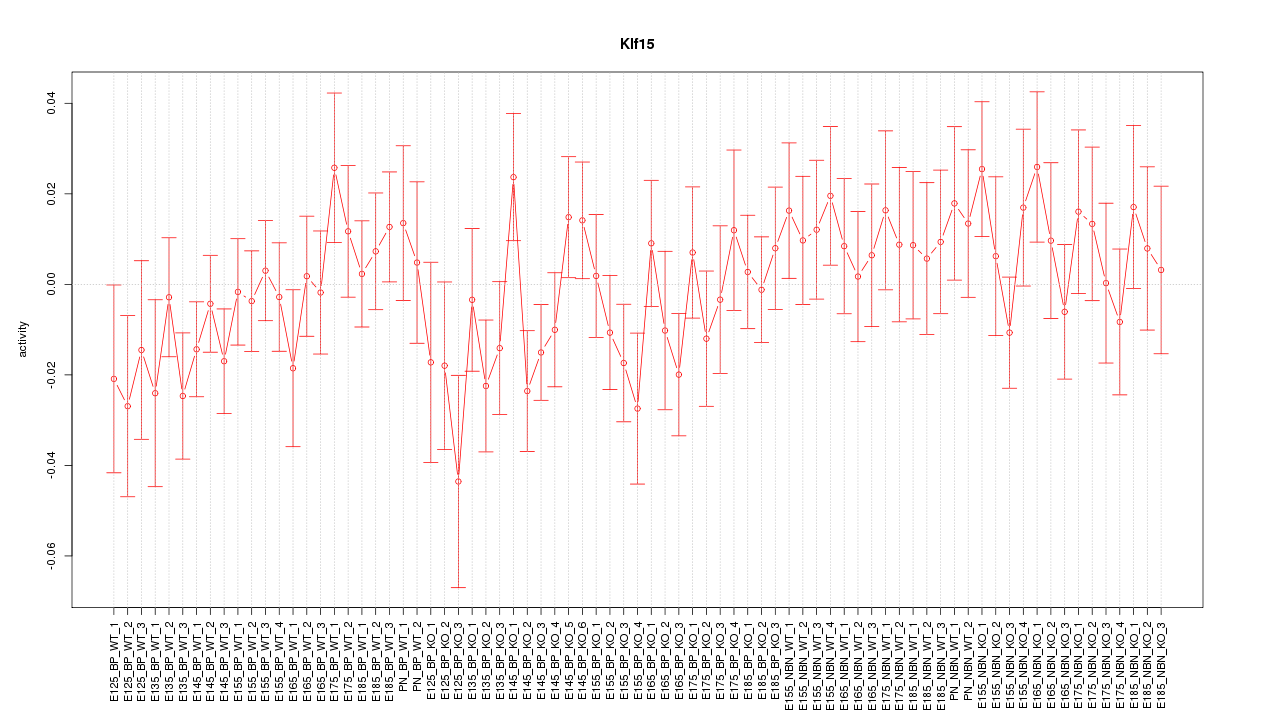
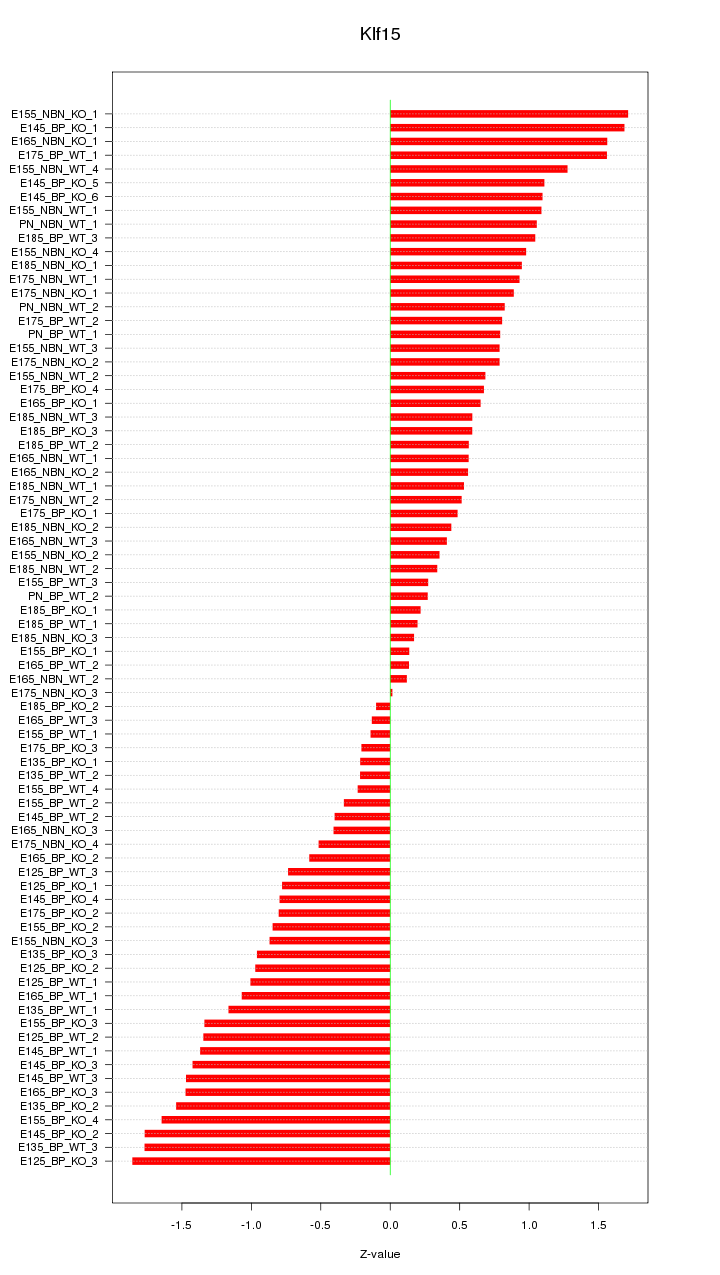
Motif ID: Klf15
Z-value: 0.931
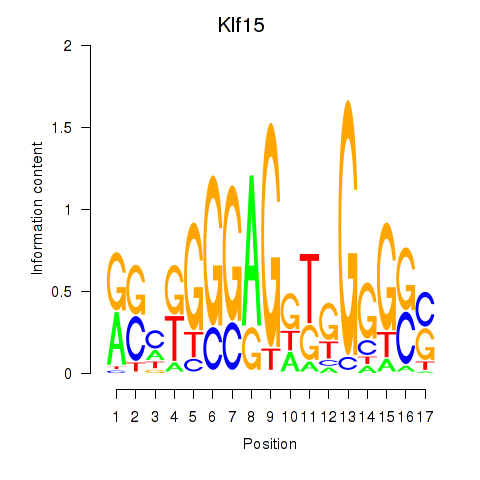
Transcription factors associated with Klf15:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Klf15 | ENSMUSG00000030087.5 | Klf15 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Klf15 | mm10_v2_chr6_+_90465287_90465304 | 0.57 | 5.1e-08 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.0 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 2.0 | 12.0 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 2.0 | 8.0 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.6 | 26.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.5 | 4.4 | GO:0060596 | mammary placode formation(GO:0060596) |
| 1.5 | 4.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 1.3 | 7.9 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 1.1 | 2.3 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 1.1 | 7.9 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 1.0 | 6.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 1.0 | 3.0 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.9 | 2.8 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.8 | 2.5 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.8 | 3.3 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.8 | 5.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.7 | 3.7 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.7 | 3.5 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.7 | 3.4 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.6 | 3.9 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.6 | 2.6 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.6 | 3.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.6 | 4.9 | GO:0061368 | maternal process involved in parturition(GO:0060137) behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.6 | 3.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.5 | 2.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.5 | 1.5 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.5 | 2.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.5 | 2.0 | GO:0021764 | amygdala development(GO:0021764) |
| 0.5 | 10.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.5 | 12.5 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.4 | 4.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.4 | 2.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.4 | 3.0 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.4 | 1.7 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.4 | 1.3 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.4 | 3.2 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.4 | 1.2 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.4 | 1.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.4 | 2.2 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.4 | 1.4 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.4 | 2.9 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 5.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.4 | 3.5 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.3 | 5.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.3 | 1.0 | GO:1902460 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.3 | 1.3 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.3 | 1.3 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.3 | 0.9 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.3 | 1.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 0.9 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.3 | 1.4 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.3 | 1.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.3 | 2.8 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.3 | 2.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.3 | 1.0 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 | 2.7 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.2 | 1.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 2.7 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.2 | 1.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 1.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 1.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 2.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 2.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.2 | 6.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 0.8 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.2 | 0.6 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.2 | 1.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 0.5 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.2 | 4.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.7 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.2 | 0.5 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.2 | 0.5 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.2 | 2.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 1.0 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) |
| 0.2 | 0.9 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 5.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 0.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 0.6 | GO:0072236 | DCT cell differentiation(GO:0072069) metanephric loop of Henle development(GO:0072236) metanephric DCT cell differentiation(GO:0072240) |
| 0.1 | 4.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 4.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 1.5 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) observational learning(GO:0098597) |
| 0.1 | 0.3 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.1 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 | 0.4 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.1 | 0.4 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 0.7 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.1 | 0.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.5 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 3.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 1.9 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.1 | 1.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.4 | GO:1903527 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 4.1 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.1 | 1.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.7 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.1 | 0.7 | GO:0006560 | proline metabolic process(GO:0006560) |
| 0.1 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.4 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.5 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 5.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 2.4 | GO:0017145 | stem cell division(GO:0017145) |
| 0.1 | 0.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 1.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 3.7 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.1 | 1.3 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 0.5 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 0.7 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.4 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 0.4 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 0.5 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 1.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.0 | 1.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.8 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 1.7 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 1.2 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.1 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 2.2 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 1.5 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 2.0 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 1.0 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 1.5 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 1.9 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.3 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 1.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 1.1 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 1.1 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.0 | 1.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.5 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 0.8 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.0 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 | 1.6 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.6 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.6 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 2.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.1 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.6 | GO:0055085 | transmembrane transport(GO:0055085) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0030934 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 1.5 | 3.0 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 1.2 | 3.7 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.8 | 4.2 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.7 | 2.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.6 | 3.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.6 | 3.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.4 | 6.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 1.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.3 | 9.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 21.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 1.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 3.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.2 | 3.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 11.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 0.5 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.1 | 1.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 3.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 10.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 2.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 2.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.5 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 5.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 5.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 6.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 10.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 2.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 3.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.7 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 5.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 5.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.4 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 3.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.4 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 4.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 6.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 9.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 2.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 2.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.5 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 4.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.4 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 7.2 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.8 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.7 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.6 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 1.5 | 4.4 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 1.1 | 10.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 1.0 | 7.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.9 | 3.7 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.9 | 3.7 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.7 | 2.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.6 | 4.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.6 | 3.4 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.6 | 3.3 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.5 | 2.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 2.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.5 | 5.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 2.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.4 | 1.7 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.4 | 4.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.4 | 2.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.4 | 9.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.4 | 2.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.4 | 2.9 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.4 | 13.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.4 | 3.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 1.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 4.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 1.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 1.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 2.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 1.7 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 11.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 1.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.3 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 6.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 0.6 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 0.6 | GO:0070736 | protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) |
| 0.2 | 2.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 2.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 12.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 1.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 5.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 4.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 3.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.1 | 2.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 3.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 7.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 4.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 1.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 2.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 3.8 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 0.7 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 2.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 2.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 5.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.7 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 3.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 1.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.8 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 2.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 2.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 1.1 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 4.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 5.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.6 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.8 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.8 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 3.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.8 | GO:0005262 | calcium channel activity(GO:0005262) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.6 | PID_S1P_S1P1_PATHWAY | S1P1 pathway |
| 0.3 | 4.4 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.3 | 12.8 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 9.4 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.3 | 4.6 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 4.9 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.1 | 11.4 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 6.2 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.9 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.1 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.8 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 6.1 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.1 | 3.9 | PID_PI3K_PLC_TRK_PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 3.1 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.3 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.4 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 0.4 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 11.9 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.0 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.0 | 1.8 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.1 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.4 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.4 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 1.0 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.3 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 3.9 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID_AVB3_INTEGRIN_PATHWAY | Integrins in angiogenesis |
| 0.0 | 1.8 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.6 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.0 | 0.4 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 1.0 | PID_P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.5 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.4 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.5 | 1.4 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.5 | 2.8 | REACTOME_COMMON_PATHWAY | Genes involved in Common Pathway |
| 0.3 | 3.3 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 6.1 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.2 | 12.8 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.2 | 1.6 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 1.1 | REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 3.8 | REACTOME_KERATAN_SULFATE_BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 9.2 | REACTOME_POST_NMDA_RECEPTOR_ACTIVATION_EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.1 | 3.4 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 4.4 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.7 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.3 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 6.6 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.9 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.0 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.2 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 2.2 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.7 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.6 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.0 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 5.3 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 1.0 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 1.2 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.5 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.0 | 2.0 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME_NFKB_ACTIVATION_THROUGH_FADD_RIP1_PATHWAY_MEDIATED_BY_CASPASE_8_AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.2 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.9 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.1 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME_ELONGATION_ARREST_AND_RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.3 | REACTOME_DNA_REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.0 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_PDGF | Genes involved in Signaling by PDGF |