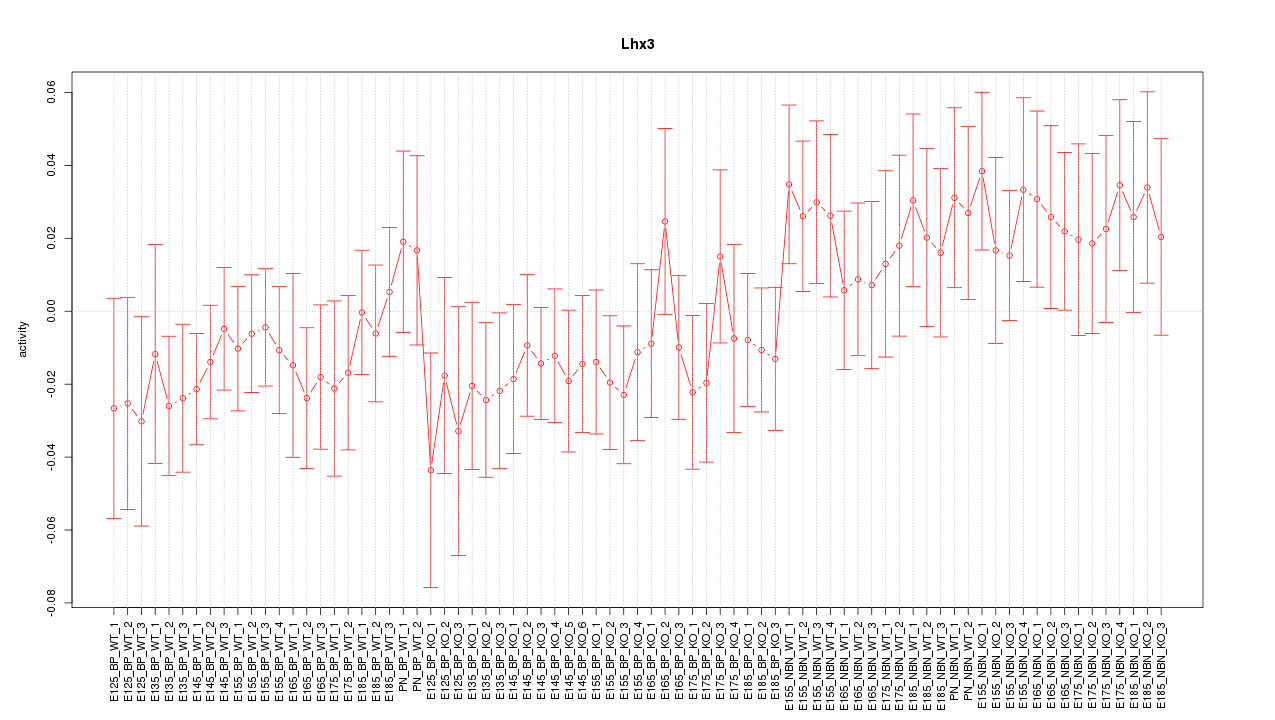
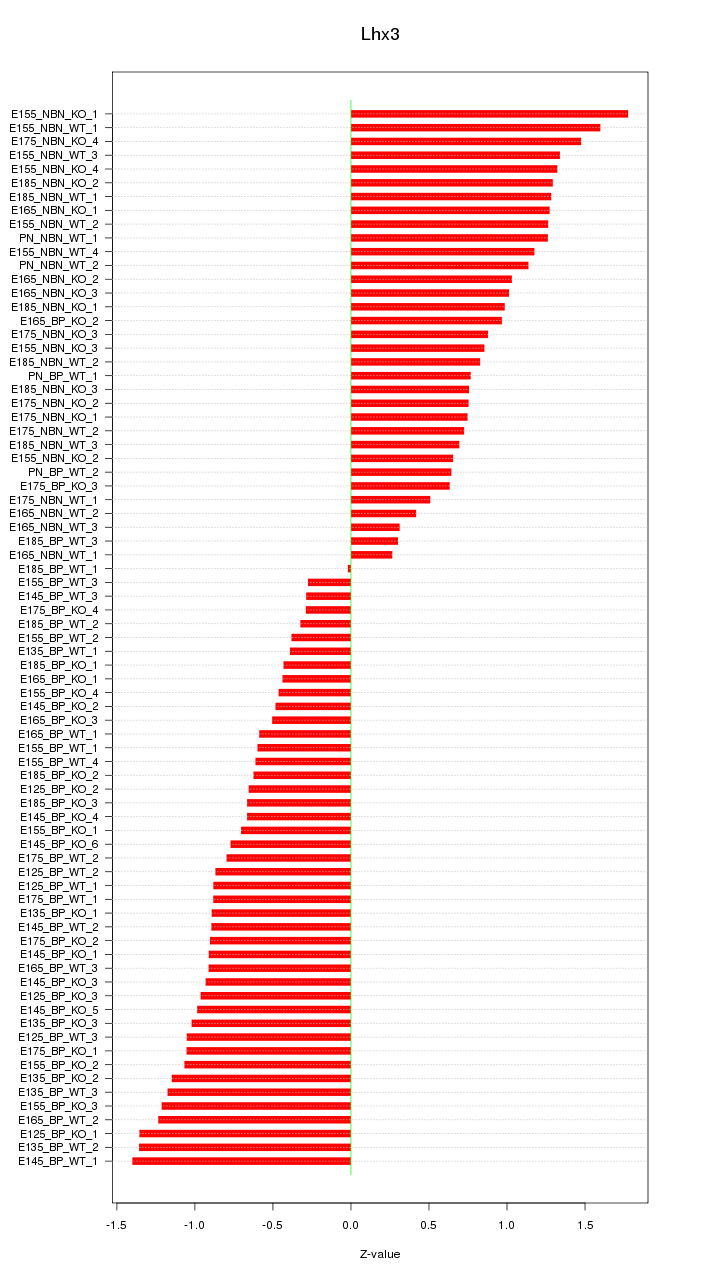
Motif ID: Lhx3
Z-value: 0.917
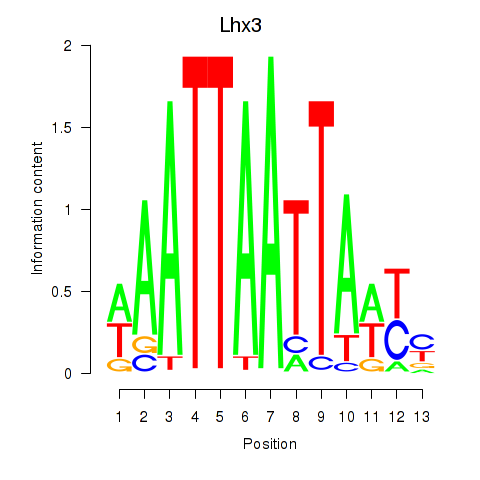
Transcription factors associated with Lhx3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Lhx3 | ENSMUSG00000026934.9 | Lhx3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 34.9 | GO:0019532 | oxalate transport(GO:0019532) |
| 2.4 | 9.7 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 1.0 | 3.0 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.9 | 8.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.9 | 2.7 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.9 | 2.7 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.8 | 3.4 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.8 | 4.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.7 | 4.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.7 | 10.7 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.7 | 2.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.6 | 6.4 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.6 | 8.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.6 | 3.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.5 | 6.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.5 | 3.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.4 | 9.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.3 | 6.3 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.2 | 0.7 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 0.9 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.2 | 3.2 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.2 | 4.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 2.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 6.0 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.4 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 1.5 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 6.1 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 2.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 3.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 6.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 4.9 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.1 | 4.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.9 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 14.1 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 6.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 2.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 2.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 2.4 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.8 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 2.0 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 14.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.0 | 4.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.7 | 2.2 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.6 | 3.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.5 | 3.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 8.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.4 | 6.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 2.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.3 | 8.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 1.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 6.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 6.5 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.5 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 7.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 29.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 6.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 6.1 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 10.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 6.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 9.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 2.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 14.9 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 3.2 | 34.9 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 2.6 | 12.9 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 1.6 | 6.4 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.8 | 4.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.8 | 4.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.7 | 2.7 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.6 | 6.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.6 | 3.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.5 | 3.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 6.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 2.2 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.3 | 8.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 4.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 6.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 3.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 8.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 6.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.0 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 8.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 3.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 2.2 | GO:0050811 | GABA-A receptor activity(GO:0004890) GABA receptor binding(GO:0050811) |
| 0.1 | 2.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 9.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 3.4 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 2.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.4 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 16.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 5.3 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 2.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 4.8 | GO:0003779 | actin binding(GO:0003779) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.4 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.2 | 8.0 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.1 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 11.4 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.6 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.7 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 4.0 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.0 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.4 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.5 | 5.3 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 34.9 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.3 | 11.4 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 2.0 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 4.1 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 6.3 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.2 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 4.1 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 3.5 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 8.1 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.5 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 2.7 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.0 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.3 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.7 | REACTOME_L1CAM_INTERACTIONS | Genes involved in L1CAM interactions |