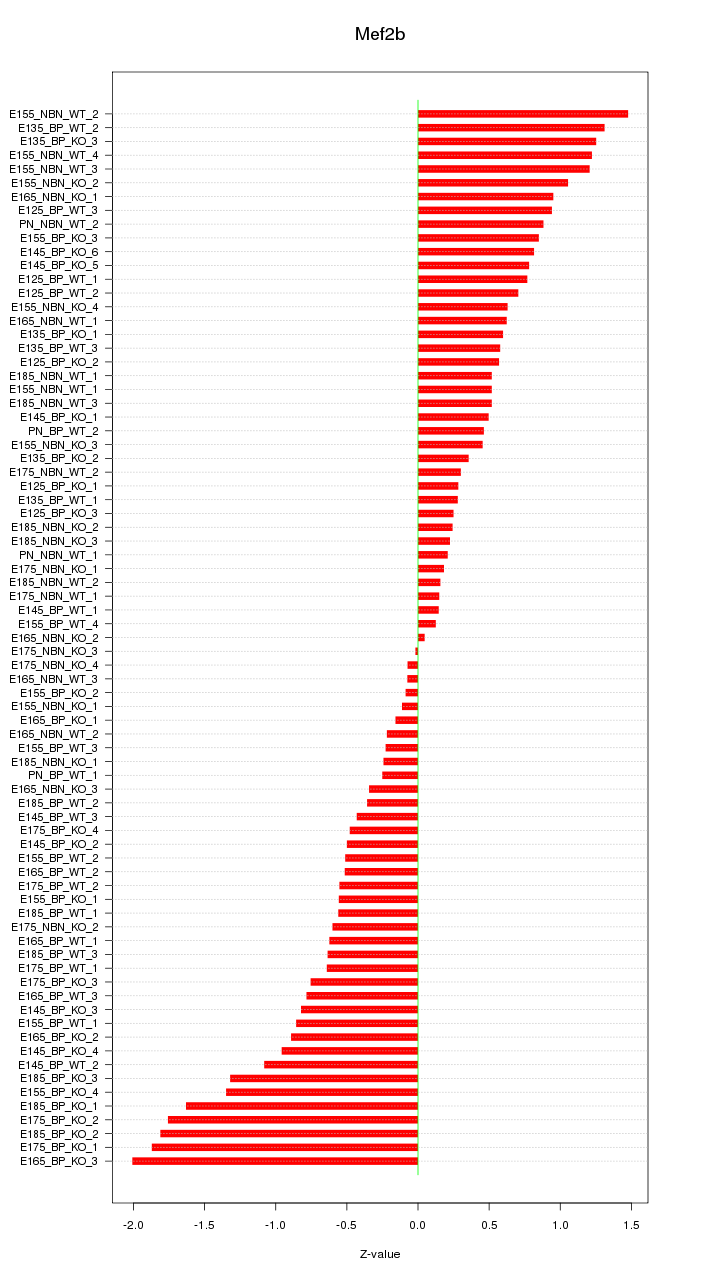
Motif ID: Mef2b
Z-value: 0.798

Transcription factors associated with Mef2b:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mef2b | ENSMUSG00000079033.3 | Mef2b |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2b | mm10_v2_chr8_+_70152754_70152781 | 0.06 | 6.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 8.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.5 | 7.3 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 1.3 | 6.3 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 1.0 | 4.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.9 | 3.6 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.8 | 2.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.7 | 3.0 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.7 | 6.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.7 | 2.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.7 | 2.0 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.7 | 3.3 | GO:0090669 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) telomerase RNA stabilization(GO:0090669) |
| 0.7 | 3.9 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.6 | 1.9 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K6-linked deubiquitination(GO:0044313) |
| 0.6 | 1.9 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.6 | 1.8 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.6 | 3.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.5 | 9.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.5 | 2.0 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.5 | 1.5 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.5 | 2.9 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.5 | 5.7 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.4 | 2.9 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.3 | 3.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.3 | 2.4 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.3 | 3.5 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.3 | 1.7 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.3 | 0.8 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.3 | 2.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 | 3.0 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.2 | 1.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.2 | 2.5 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 1.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 2.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 0.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 1.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.6 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 1.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.7 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.4 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.4 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.1 | 2.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.5 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 0.5 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell differentiation(GO:0010668) |
| 0.1 | 7.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 5.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.6 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.5 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.9 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.3 | GO:0006706 | steroid catabolic process(GO:0006706) |
| 0.1 | 0.7 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 1.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.4 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.3 | GO:0090282 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.1 | 1.9 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.0 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 2.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.7 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 0.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.3 | GO:0098535 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 0.0 | 1.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 2.2 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.6 | GO:0019369 | drug metabolic process(GO:0017144) arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 1.1 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 2.7 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.6 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.8 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 3.6 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 1.4 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.8 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.0 | 3.5 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.5 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.4 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 1.8 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.0 | 1.2 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 4.1 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 3.8 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 1.3 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0045346 | regulation of MHC class II biosynthetic process(GO:0045346) negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 3.4 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 1.2 | 3.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.8 | 3.3 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.6 | 2.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.6 | 2.2 | GO:0090537 | CERF complex(GO:0090537) |
| 0.4 | 3.0 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 2.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 2.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.2 | 3.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 0.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 3.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 4.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 2.0 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 7.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 1.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 6.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 10.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 3.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 1.0 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 2.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 5.0 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 12.4 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 4.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 6.3 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 10.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.2 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 3.0 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0036126 | sperm flagellum(GO:0036126) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.5 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 1.4 | 9.5 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.1 | 3.4 | GO:0016160 | alpha-amylase activity(GO:0004556) amylase activity(GO:0016160) |
| 0.8 | 3.0 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.7 | 2.2 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.7 | 3.6 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.7 | 2.9 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.7 | 2.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.6 | 3.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.5 | 3.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.5 | 5.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.4 | 1.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.4 | 1.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 3.0 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.2 | 7.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.2 | 0.6 | GO:0004534 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 0.6 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.2 | 0.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.2 | 2.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 1.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 2.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 1.9 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 3.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 3.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.3 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 1.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 2.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.3 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.1 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 2.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 5.0 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.1 | 2.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 2.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 6.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.4 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 2.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 2.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.6 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 2.3 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.8 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 2.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 11.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.6 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.6 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.9 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 7.8 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 4.1 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 7.3 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 3.6 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 1.9 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.1 | 3.0 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.1 | 3.9 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.8 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.5 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 5.6 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID_INTEGRIN_CS_PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.5 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.3 | 8.7 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 3.6 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.4 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 3.6 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.2 | 5.5 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.6 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.7 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 3.5 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.9 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.8 | REACTOME_THE_ROLE_OF_NEF_IN_HIV1_REPLICATION_AND_DISEASE_PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.4 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.1 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.4 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.5 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.8 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.7 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.9 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME_PYRUVATE_METABOLISM_AND_CITRIC_ACID_TCA_CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 2.5 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME_TELOMERE_MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 0.4 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.6 | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |