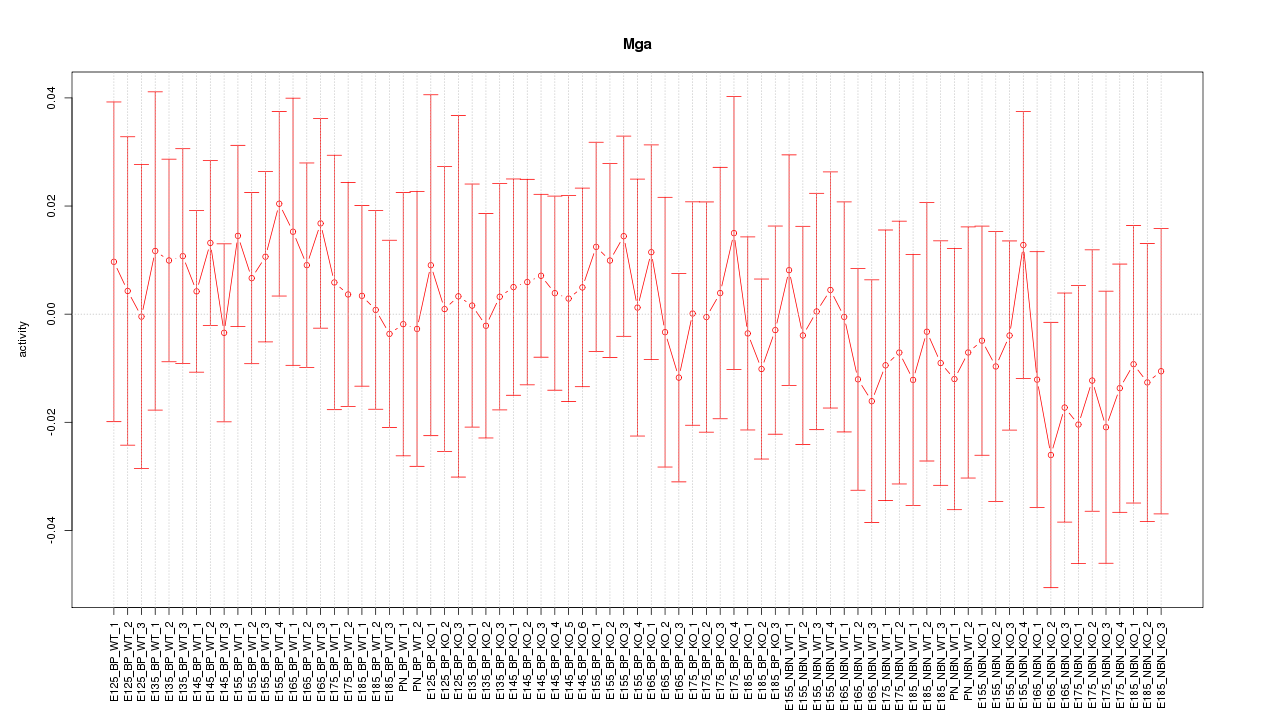
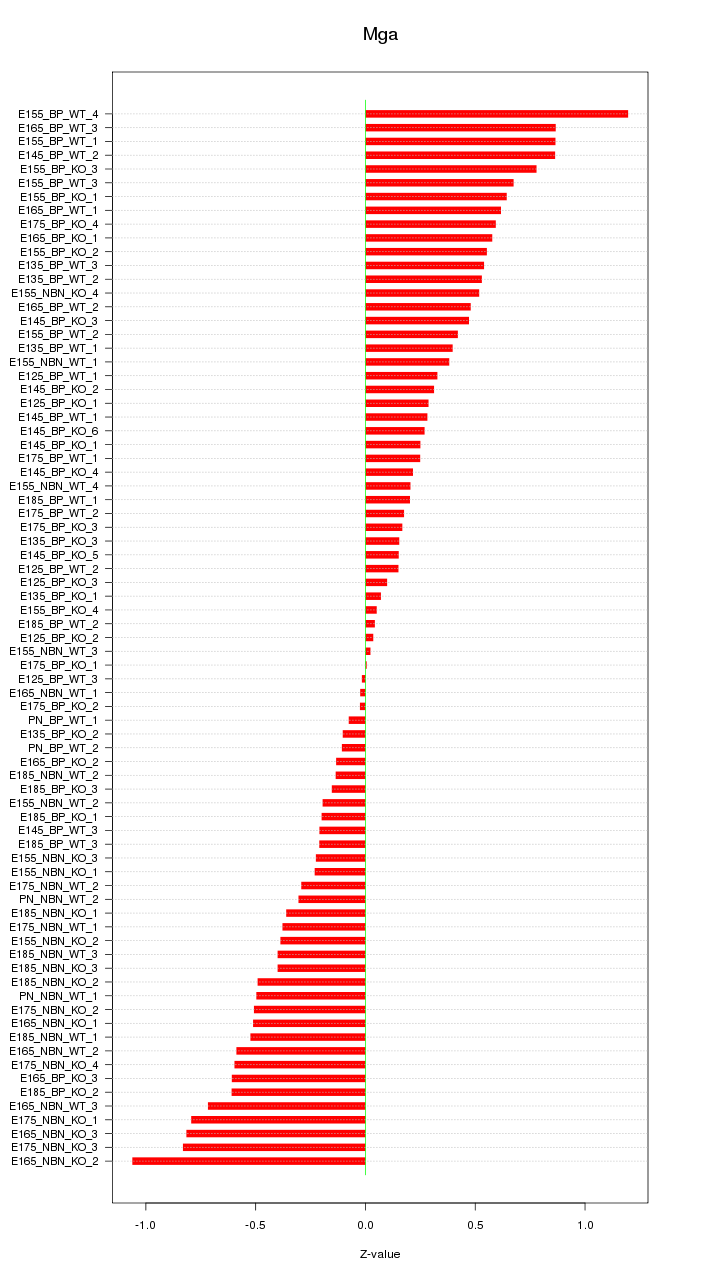
Motif ID: Mga
Z-value: 0.467
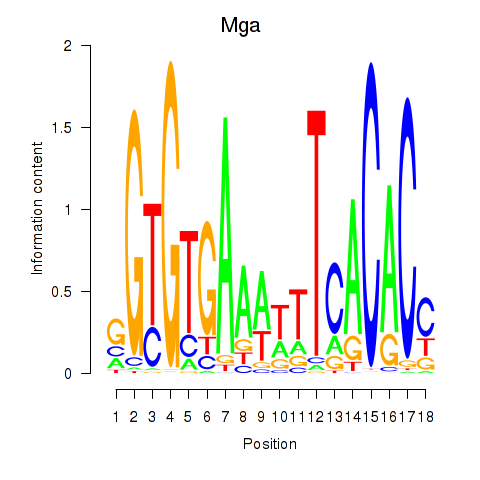
Transcription factors associated with Mga:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mga | ENSMUSG00000033943.9 | Mga |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mga | mm10_v2_chr2_+_119897212_119897305 | 0.32 | 5.2e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.4 | 1.7 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.4 | 4.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 0.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.3 | 1.3 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.2 | 1.6 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.2 | 1.9 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 4.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.4 | GO:0006592 | ornithine biosynthetic process(GO:0006592) |
| 0.1 | 2.5 | GO:0071803 | keratinocyte development(GO:0003334) positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.4 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) |
| 0.1 | 0.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.8 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.6 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 5.2 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.1 | 0.3 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.3 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.7 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 1.0 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.5 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 2.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 2.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 2.3 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.6 | GO:0060612 | adipose tissue development(GO:0060612) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 4.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 0.7 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 2.0 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.1 | 2.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.0 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 2.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 1.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 1.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.4 | GO:0017084 | glutamate 5-kinase activity(GO:0004349) glutamate-5-semialdehyde dehydrogenase activity(GO:0004350) delta1-pyrroline-5-carboxylate synthetase activity(GO:0017084) amino acid kinase activity(GO:0019202) |
| 0.1 | 1.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.9 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.7 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 1.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 0.4 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 1.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 4.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 2.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 4.2 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.7 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.9 | PID_CD8_TCR_PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 2.0 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 0.0 | 1.8 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID_ARF6_PATHWAY | Arf6 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.6 | REACTOME_PROCESSIVE_SYNTHESIS_ON_THE_LAGGING_STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.1 | 1.6 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |