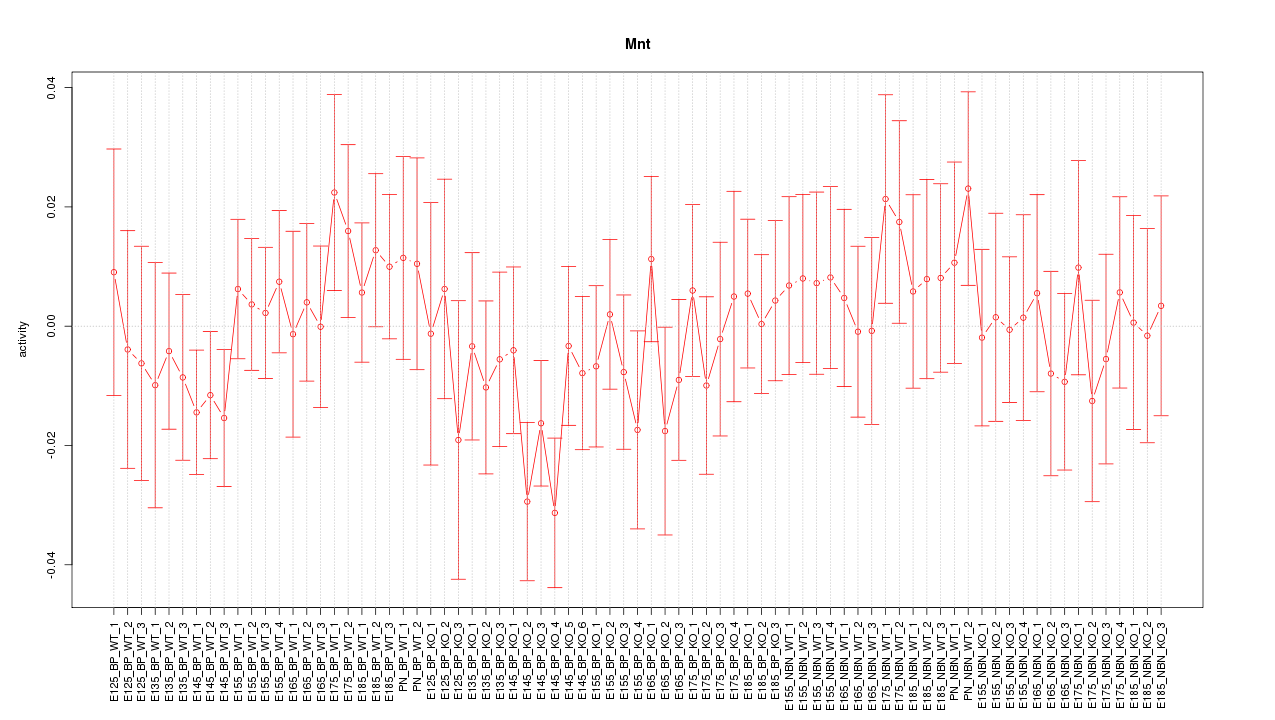
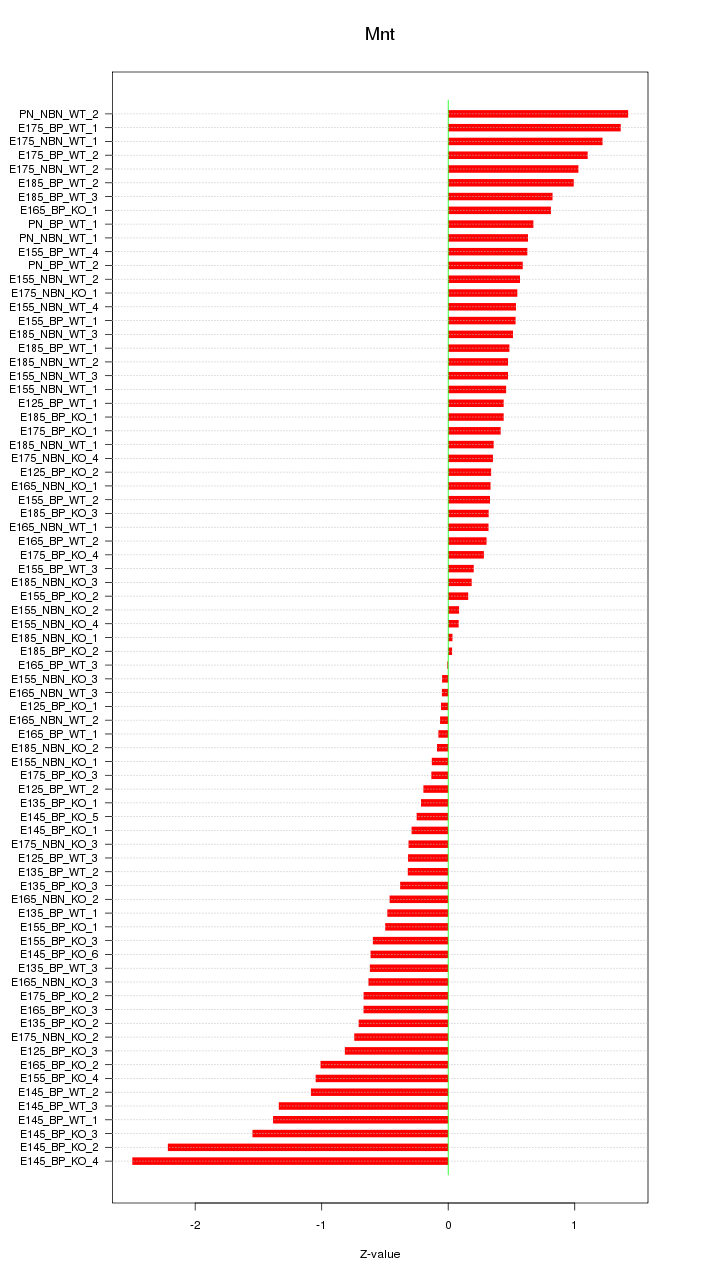
Motif ID: Mnt
Z-value: 0.735

Transcription factors associated with Mnt:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Mnt | ENSMUSG00000000282.6 | Mnt |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mnt | mm10_v2_chr11_+_74830920_74831005 | -0.29 | 1.1e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 1.8 | 5.4 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.8 | 12.3 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.6 | 4.8 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 1.3 | 3.9 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.1 | 3.2 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 1.0 | 2.9 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.9 | 2.8 | GO:1902022 | L-lysine transport(GO:1902022) |
| 0.9 | 3.6 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.9 | 4.4 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.9 | 3.5 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.8 | 4.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.7 | 2.2 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.7 | 2.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.7 | 2.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.7 | 2.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.7 | 2.8 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.7 | 3.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.7 | 4.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.7 | 1.4 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.7 | 2.7 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 0.7 | 2.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.7 | 2.0 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.6 | 2.6 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.6 | 3.4 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.6 | 2.8 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.5 | 1.1 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.5 | 3.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.5 | 2.6 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.5 | 2.5 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.5 | 1.5 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.5 | 2.0 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.5 | 1.4 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.5 | 0.9 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.5 | 1.8 | GO:0006272 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.4 | 2.2 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 | 1.2 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.4 | 1.6 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 | 3.8 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.4 | 1.5 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.4 | 1.5 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.4 | 1.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.4 | 1.8 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.4 | 1.8 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.4 | 1.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 1.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.4 | 1.4 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.4 | 1.1 | GO:0045359 | positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.3 | 1.3 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.3 | 1.0 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.3 | 2.5 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.3 | 5.7 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 1.2 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.3 | 0.9 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.3 | 3.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 1.2 | GO:0010046 | arginine biosynthetic process(GO:0006526) response to mycotoxin(GO:0010046) |
| 0.3 | 1.2 | GO:2001245 | phospholipid homeostasis(GO:0055091) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.3 | 1.2 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.3 | 2.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.3 | 1.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.3 | 2.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.3 | 2.4 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.3 | 0.8 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.3 | 1.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 2.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 1.8 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.3 | 2.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.3 | 0.8 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.3 | 1.3 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.2 | 1.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 1.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.2 | 2.7 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 1.7 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 0.9 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.2 | 0.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.2 | 1.6 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 1.8 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.2 | 1.1 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.2 | 2.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.2 | 0.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 | 0.6 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 0.6 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.2 | 1.7 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 1.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 0.8 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.2 | 1.0 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.2 | 0.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 0.6 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 1.4 | GO:0072015 | IMP salvage(GO:0032264) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 0.4 | GO:0021586 | pons maturation(GO:0021586) |
| 0.2 | 1.2 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.2 | 0.6 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.2 | 1.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 2.7 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.2 | 1.2 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 4.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 | 1.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 2.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 1.0 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 0.7 | GO:0046655 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) folic acid metabolic process(GO:0046655) |
| 0.2 | 1.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 1.5 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.2 | 0.6 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.2 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 1.9 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.2 | 0.9 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 | 5.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 1.7 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 0.9 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.2 | 2.3 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 1.2 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 0.8 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 0.1 | 2.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 2.7 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 1.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.4 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 1.5 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.9 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.1 | 0.8 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 0.6 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 3.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 1.1 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.1 | 0.5 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 1.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 3.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 1.6 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 1.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.0 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 | 3.5 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 1.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.9 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.7 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.4 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.3 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.1 | 1.2 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.3 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 1.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.5 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.6 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.8 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 1.1 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 0.3 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 2.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 2.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.7 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 3.7 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 3.6 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.1 | 0.9 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.6 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.5 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.1 | 0.3 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.8 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.1 | 0.8 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 1.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 1.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 1.0 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.3 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 3.4 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 0.2 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.1 | 0.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.2 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 0.2 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 3.4 | GO:0043297 | apical junction assembly(GO:0043297) |
| 0.1 | 3.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 1.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.4 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.1 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.4 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 2.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 2.0 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.1 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 2.0 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.6 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 1.1 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.5 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) proton-transporting two-sector ATPase complex assembly(GO:0070071) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.2 | GO:1902219 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.3 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 1.0 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 0.7 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.2 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 1.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.3 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.6 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 6.9 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 1.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 2.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.5 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.4 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.7 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.0 | 1.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 2.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 1.1 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.0 | 0.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.1 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 2.3 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.9 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 1.8 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 1.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.5 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.4 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.6 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.2 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.4 | GO:0010043 | response to zinc ion(GO:0010043) |
| 0.0 | 1.5 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 1.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.5 | GO:0007292 | female gamete generation(GO:0007292) |
| 0.0 | 1.0 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 1.1 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.2 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 1.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 1.4 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 1.3 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 1.6 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.4 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.1 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0060486 | Clara cell differentiation(GO:0060486) epithelial cell proliferation involved in lung morphogenesis(GO:0060502) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 1.3 | GO:0000209 | protein polyubiquitination(GO:0000209) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.1 | 5.4 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.6 | 1.9 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.6 | 2.9 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.6 | 1.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.6 | 2.2 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.5 | 3.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.5 | 2.0 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.5 | 1.4 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.4 | 7.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 2.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 1.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.4 | 1.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.4 | 3.8 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.4 | 1.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 2.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 1.8 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 2.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.3 | 2.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 0.9 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.3 | 1.5 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.3 | 2.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 5.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.3 | 3.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.3 | 1.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.3 | 1.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 1.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 1.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.2 | 2.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 0.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 0.7 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 1.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 0.6 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 2.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 1.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 12.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 0.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 1.8 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.2 | 2.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 0.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 0.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 3.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.9 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 1.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 2.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 4.1 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.3 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.4 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 1.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 2.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.6 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 1.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.3 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.7 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 2.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 4.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 1.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 4.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.9 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.7 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 2.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 3.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 1.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.2 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 3.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 1.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.5 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 4.6 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 3.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.2 | GO:0099568 | cytoplasmic region(GO:0099568) |
| 0.0 | 0.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 12.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.2 | 4.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 1.1 | 3.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 1.0 | 4.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.9 | 5.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.9 | 2.6 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.7 | 2.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.7 | 4.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.6 | 3.9 | GO:0043426 | MRF binding(GO:0043426) |
| 0.6 | 2.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.5 | 3.0 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.5 | 1.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.5 | 1.4 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.5 | 4.5 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.4 | 1.3 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.4 | 2.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.4 | 1.7 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.4 | 1.2 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.4 | 3.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.4 | 1.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.4 | 1.5 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.4 | 1.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 1.7 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 7.4 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.3 | 2.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 5.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.3 | 2.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.3 | 5.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 2.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 2.3 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.3 | 0.8 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.3 | 1.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 1.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 0.8 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.2 | 2.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 2.0 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 1.7 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.2 | 1.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 1.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 1.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 0.7 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 0.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 1.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 0.8 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.2 | 1.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 2.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 1.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 4.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 1.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 1.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 5.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 5.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 3.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 0.9 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 0.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 2.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.2 | 2.0 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 0.7 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.2 | 1.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 0.9 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 0.8 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 1.0 | GO:0005047 | signal recognition particle binding(GO:0005047) 7S RNA binding(GO:0008312) |
| 0.1 | 1.6 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.4 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.1 | 0.4 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.1 | 0.4 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.1 | 1.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 3.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 6.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.4 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 3.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.5 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.1 | 1.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 1.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 1.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 3.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 3.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 2.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 4.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 2.2 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.3 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 1.2 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 0.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 1.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 1.0 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 1.6 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.6 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.7 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.3 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 1.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.9 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 2.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.2 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 0.1 | 2.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 1.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 3.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.7 | GO:0090079 | translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.1 | 5.1 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.9 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.0 | 1.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 3.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 3.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.5 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) DNA polymerase activity(GO:0034061) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.1 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.4 | 1.4 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 12.6 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.3 | 5.2 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.2 | 5.0 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.2 | 3.5 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.9 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 6.9 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.1 | 2.9 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.3 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 0.9 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.1 | 2.7 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.4 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 4.0 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 2.4 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 1.4 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.0 | 0.9 | PID_S1P_S1P3_PATHWAY | S1P3 pathway |
| 0.0 | 1.8 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.5 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.9 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.7 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.5 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.4 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 5.8 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.1 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.3 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.0 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.0 | 1.6 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.7 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.8 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID_IFNG_PATHWAY | IFN-gamma pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 5.5 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 12.1 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.3 | 3.2 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 2.7 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 1.8 | REACTOME_E2F_ENABLED_INHIBITION_OF_PRE_REPLICATION_COMPLEX_FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 5.5 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 3.6 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 2.2 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.8 | REACTOME_POL_SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 0.5 | REACTOME_INFLUENZA_LIFE_CYCLE | Genes involved in Influenza Life Cycle |
| 0.1 | 1.8 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 5.6 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.6 | REACTOME_TRANSFERRIN_ENDOCYTOSIS_AND_RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.3 | REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 4.4 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.3 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 6.4 | REACTOME_HEPARAN_SULFATE_HEPARIN_HS_GAG_METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.1 | 2.7 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.1 | REACTOME_ACTIVATION_OF_IRF3_IRF7_MEDIATED_BY_TBK1_IKK_EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 2.1 | REACTOME_CA_DEPENDENT_EVENTS | Genes involved in Ca-dependent events |
| 0.1 | 1.5 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.4 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.9 | REACTOME_P53_DEPENDENT_G1_DNA_DAMAGE_RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.1 | 1.8 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.5 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 3.0 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.1 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.2 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.0 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.4 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.7 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 3.2 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.4 | REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.7 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.0 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.9 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.6 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 2.6 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.1 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 1.8 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME_ACETYLCHOLINE_BINDING_AND_DOWNSTREAM_EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.0 | 1.1 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.1 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.1 | REACTOME_VIF_MEDIATED_DEGRADATION_OF_APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 1.2 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.4 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |