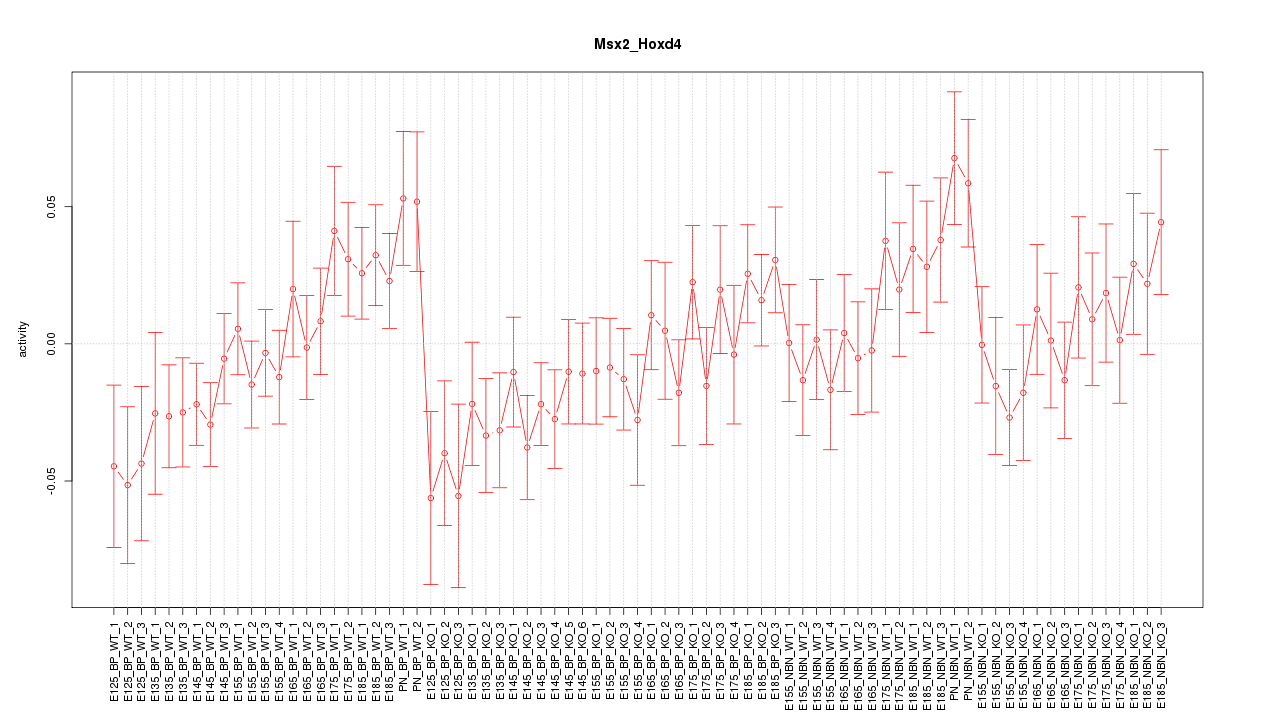
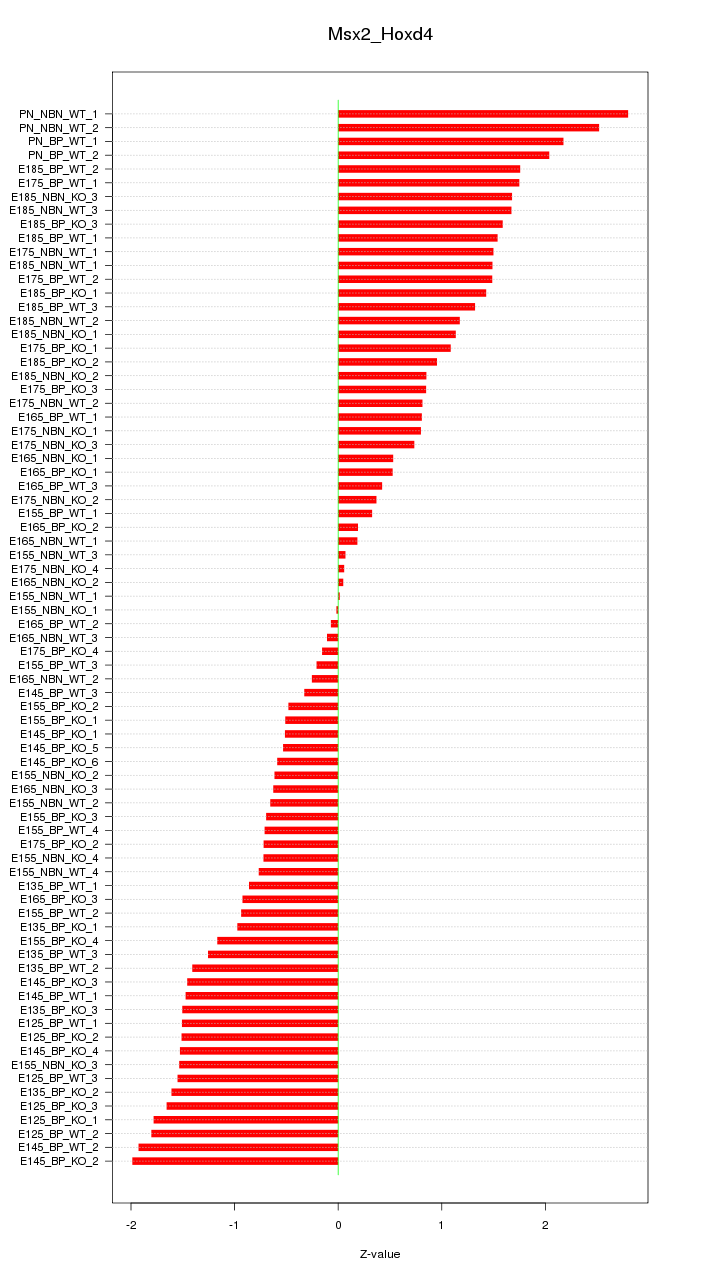
Motif ID: Msx2_Hoxd4
Z-value: 1.203
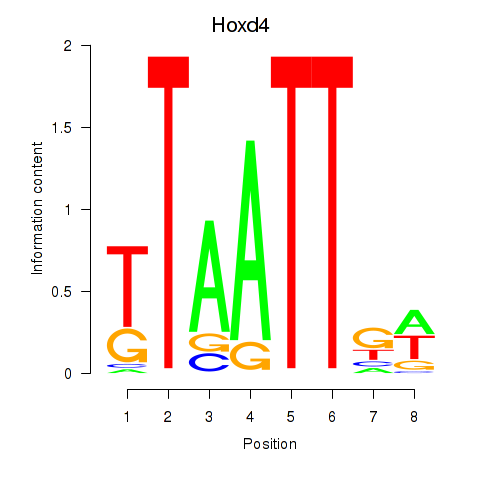
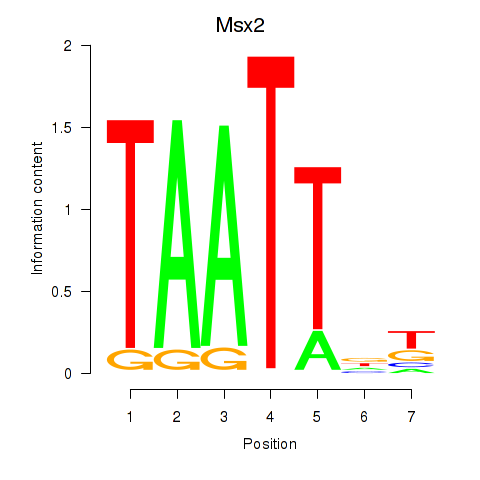
Transcription factors associated with Msx2_Hoxd4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Msx2 | ENSMUSG00000021469.8 | Msx2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Msx2 | mm10_v2_chr13_-_53473074_53473074 | -0.25 | 3.1e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.2 | 27.5 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 7.7 | 23.2 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 2.4 | 9.7 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 2.2 | 6.5 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 1.2 | 4.9 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 1.1 | 3.4 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.1 | 3.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 1.0 | 3.8 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.8 | 13.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.7 | 15.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.7 | 4.1 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.6 | 2.9 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.5 | 9.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.5 | 1.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.5 | 2.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.5 | 4.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.5 | 1.8 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.4 | 12.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.4 | 17.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.4 | 4.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.4 | 1.8 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.3 | 3.7 | GO:0071455 | cellular response to hyperoxia(GO:0071455) |
| 0.3 | 0.9 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.3 | 2.9 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.3 | 0.9 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.3 | 1.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.3 | 21.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.3 | 7.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.2 | 5.6 | GO:0030903 | notochord development(GO:0030903) |
| 0.2 | 2.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 2.7 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.2 | 6.2 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.2 | 3.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 0.5 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 1.3 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.2 | 0.6 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.2 | 1.8 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.2 | 3.4 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.2 | 1.0 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.5 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.2 | 1.7 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 4.6 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 2.2 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.1 | 2.5 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.1 | 0.8 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 2.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 3.7 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 0.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 1.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 3.9 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 7.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 0.9 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 2.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 13.7 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 0.4 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 10.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 0.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 1.7 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.0 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 2.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.7 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.7 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 | 1.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 2.9 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 2.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0021972 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 | 1.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 2.3 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.8 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 1.3 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 3.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 10.3 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.3 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.6 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.4 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.2 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.4 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.9 | 7.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.3 | 13.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 2.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.4 | 3.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 1.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 4.9 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 3.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 25.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 4.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 0.7 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 4.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 1.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.7 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 1.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 16.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 1.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 3.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.5 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.1 | 1.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.9 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 3.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 12.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 5.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 18.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 4.4 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 1.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.6 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.0 | 40.4 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 2.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 6.3 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 23.2 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 2.3 | 27.5 | GO:0050693 | LBD domain binding(GO:0050693) |
| 1.6 | 12.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 1.5 | 8.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.1 | 3.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.9 | 6.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.9 | 2.7 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.9 | 2.7 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.6 | 12.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.6 | 4.9 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.6 | 2.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.6 | 9.7 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.5 | 1.5 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.5 | 23.8 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.4 | 2.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 3.8 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.3 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 7.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 10.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 0.5 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 1.5 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.2 | 0.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 2.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 5.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 7.5 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 3.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 2.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 4.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 2.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 17.1 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 1.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 2.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 2.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 36.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 4.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 1.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 2.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.5 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 11.2 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.6 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 1.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 5.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 6.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 23.2 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.6 | 23.8 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.2 | 7.5 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.2 | 2.9 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 3.7 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 6.5 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 3.7 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.1 | 5.0 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.1 | 2.9 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 2.7 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 11.2 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.1 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 3.1 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.3 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 2.0 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.9 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.6 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.0 | 1.1 | PID_NFAT_3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.8 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.0 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.5 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.4 | 7.3 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 11.7 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 12.9 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 2.3 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 4.9 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 3.9 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 3.7 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 24.6 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.3 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 3.1 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.9 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 4.0 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.8 | REACTOME_INTEGRIN_ALPHAIIB_BETA3_SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 1.0 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 1.4 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.5 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 2.9 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.6 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |