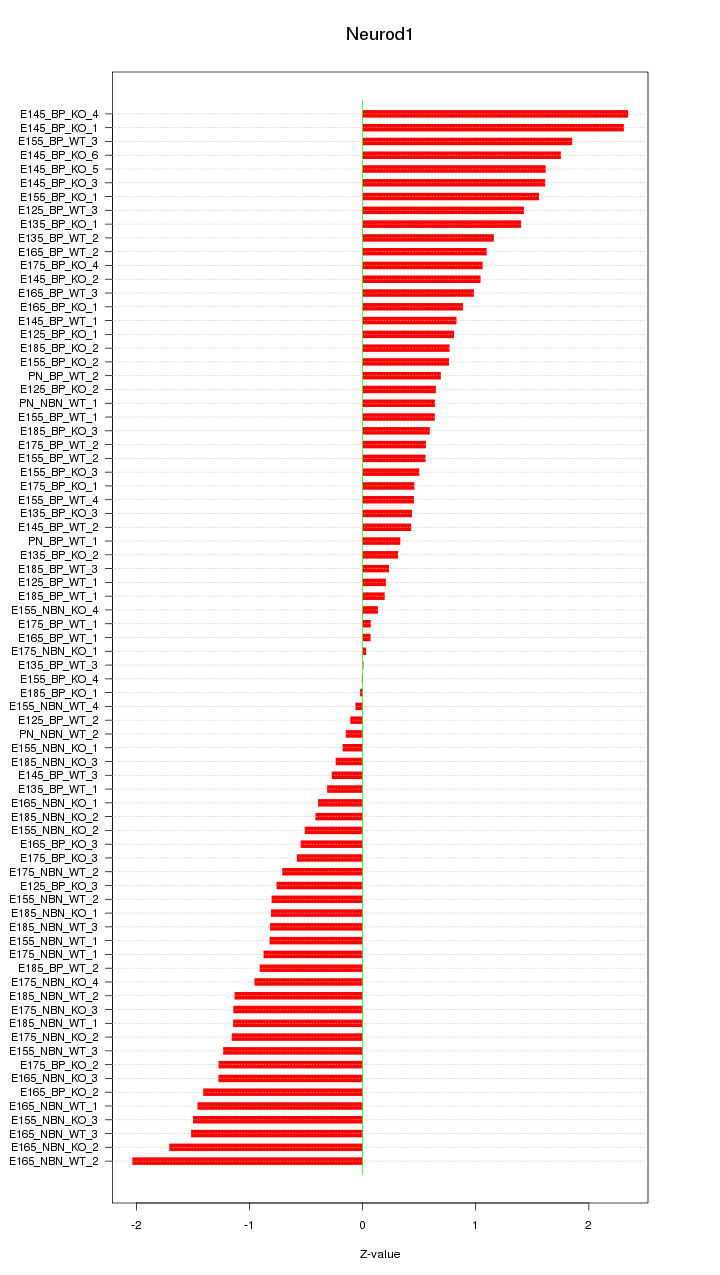
Motif ID: Neurod1
Z-value: 0.991

Transcription factors associated with Neurod1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Neurod1 | ENSMUSG00000034701.9 | Neurod1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Neurod1 | mm10_v2_chr2_-_79456750_79456761 | 0.66 | 8.3e-11 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.0 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 1.6 | 19.0 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 1.5 | 9.1 | GO:0003383 | apical constriction(GO:0003383) |
| 1.5 | 7.5 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.2 | 11.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 1.2 | 4.9 | GO:0060032 | notochord regression(GO:0060032) |
| 1.2 | 10.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 1.1 | 3.3 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.1 | 4.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 1.0 | 8.3 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 1.0 | 4.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.8 | 7.9 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.8 | 6.9 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.7 | 3.7 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.7 | 7.3 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 0.6 | 1.9 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.6 | 7.6 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.6 | 3.5 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.6 | 2.8 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.6 | 2.8 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.5 | 1.6 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.5 | 1.5 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.5 | 5.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.5 | 1.5 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.5 | 1.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.4 | 1.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.4 | 2.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.4 | 3.2 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 0.4 | 6.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 2.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 5.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.4 | 1.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.3 | 3.1 | GO:0099624 | atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.3 | 1.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.3 | 2.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 2.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.3 | 3.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.3 | 1.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.3 | 2.2 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.3 | 2.7 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 1.5 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 1.2 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.2 | 0.7 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.2 | 2.8 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.2 | 2.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 0.7 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 0.9 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 | 3.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 1.7 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 0.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.0 | GO:0071472 | cellular response to salt stress(GO:0071472) |
| 0.2 | 7.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 3.7 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 0.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 1.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 0.7 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.2 | 0.7 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 1.8 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.2 | 1.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 3.2 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.2 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 1.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 1.8 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 6.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.6 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 5.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.0 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 1.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 11.2 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 1.9 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 2.4 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.1 | 0.6 | GO:0032493 | response to bacterial lipoprotein(GO:0032493) |
| 0.1 | 2.8 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 0.1 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 7.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.5 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.1 | 1.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.1 | 3.6 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 0.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.8 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 3.0 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 1.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 2.6 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 0.3 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 1.0 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 2.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 2.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 3.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.6 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 7.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 6.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 1.3 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 | 0.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 2.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 6.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.1 | GO:0008610 | lipid biosynthetic process(GO:0008610) |
| 0.0 | 1.1 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 0.7 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 5.7 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 2.5 | GO:0051782 | negative regulation of cell division(GO:0051782) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.5 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 1.0 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 6.0 | GO:0007162 | negative regulation of cell adhesion(GO:0007162) |
| 0.0 | 2.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 3.6 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.9 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 1.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 2.6 | GO:0048515 | spermatid differentiation(GO:0048515) |
| 0.0 | 0.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 1.2 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 1.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 1.6 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 1.4 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.8 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.3 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 1.8 | GO:0010506 | regulation of autophagy(GO:0010506) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 1.7 | 6.9 | GO:0090537 | CERF complex(GO:0090537) |
| 1.0 | 15.0 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.8 | 6.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.4 | 4.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.3 | 3.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.3 | 1.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.3 | 1.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.3 | 3.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.2 | 11.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 3.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 2.7 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 13.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 3.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 2.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 1.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.2 | 14.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 24.3 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 1.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 6.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 9.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 1.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 5.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.4 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 1.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 3.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 4.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 8.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 9.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 2.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 2.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 8.1 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.2 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 3.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 3.3 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 6.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.2 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 6.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.6 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 3.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 2.0 | 6.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 1.4 | 4.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.3 | 4.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 1.3 | 3.8 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.1 | 8.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.1 | 3.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.9 | 2.8 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.9 | 17.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.6 | 1.9 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.5 | 2.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.5 | 1.5 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.4 | 2.6 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.4 | 2.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.4 | 3.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.4 | 2.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.4 | 2.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.3 | 3.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 7.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 2.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 11.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 2.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.3 | 4.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 4.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 1.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 2.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.3 | 3.3 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.3 | 4.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 1.5 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 1.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 0.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 2.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 7.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 1.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 5.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 3.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 3.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.9 | GO:0044020 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.2 | 1.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 6.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 1.9 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 9.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 1.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 1.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 3.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 3.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 3.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 4.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 6.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.7 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.0 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 1.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.7 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 4.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 6.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 2.8 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 1.9 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 16.1 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 0.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 12.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 2.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.5 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.4 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 7.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 8.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 3.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 3.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 2.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 1.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 2.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 4.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.7 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.2 | 10.4 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.2 | 5.1 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 7.3 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.2 | 3.4 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 3.5 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 6.2 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.1 | 4.0 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 4.8 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.0 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 6.0 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 2.7 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 5.9 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.3 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 3.6 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 5.3 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 6.0 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.1 | 10.7 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 6.8 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 3.2 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.8 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.1 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.3 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 3.3 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 17.0 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.6 | 11.1 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.3 | 15.9 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.3 | 7.9 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.3 | 6.0 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 4.0 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 3.1 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 3.4 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 2.5 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 8.9 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 2.1 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.2 | REACTOME_PRE_NOTCH_EXPRESSION_AND_PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 0.8 | REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 3.2 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.6 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.2 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 6.7 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.3 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.1 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 4.6 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.1 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 6.3 | REACTOME_CELL_SURFACE_INTERACTIONS_AT_THE_VASCULAR_WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 5.4 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.7 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 5.2 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.9 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 5.5 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.6 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 10.0 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.7 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.5 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.7 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.8 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.9 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |