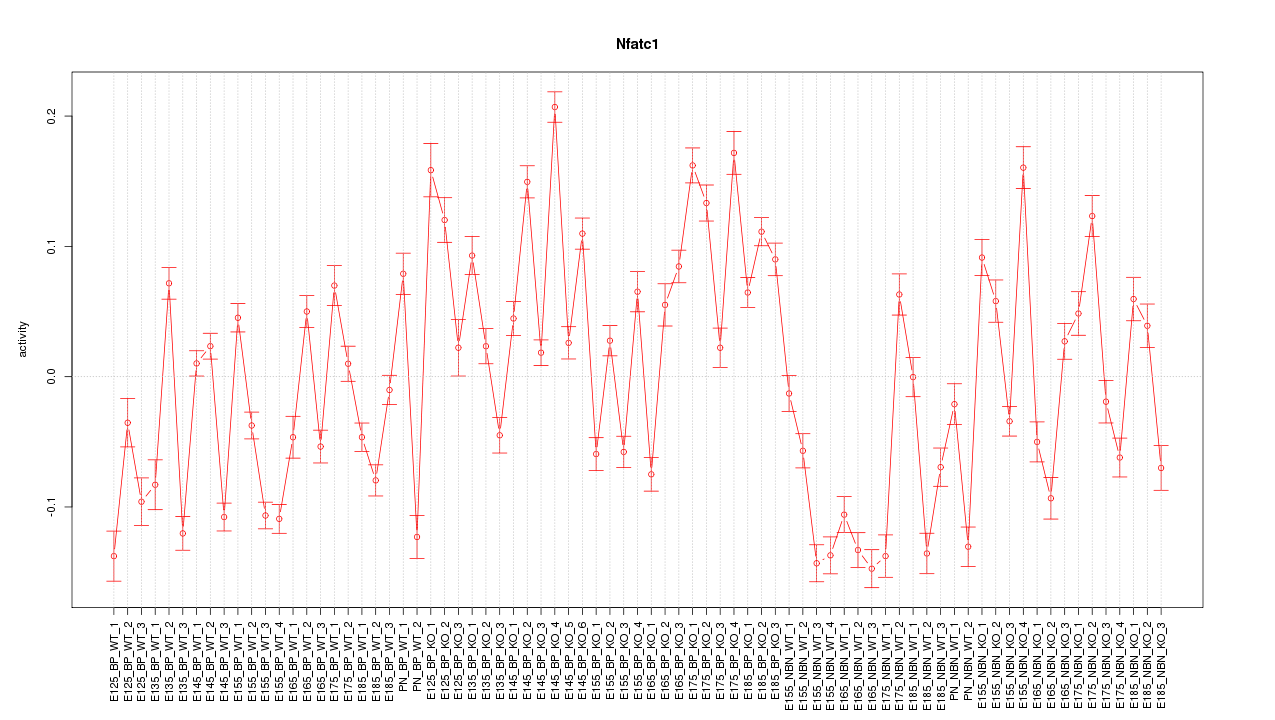
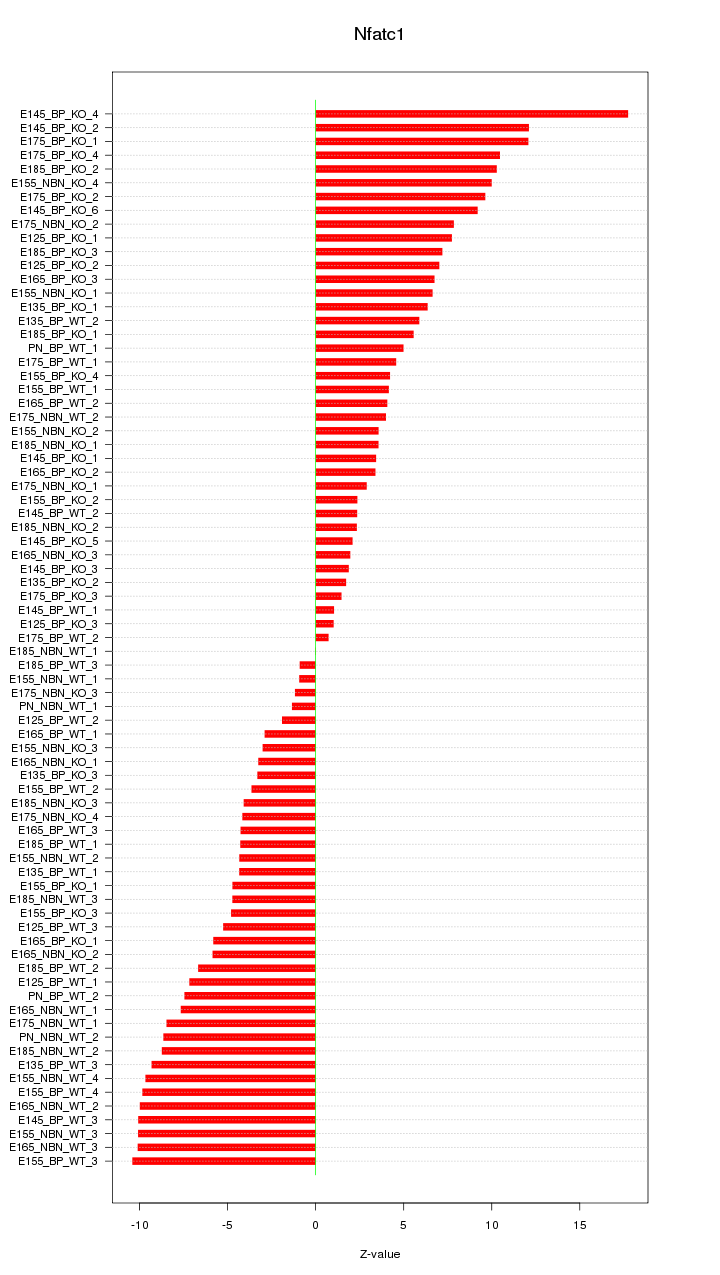
Motif ID: Nfatc1
Z-value: 6.529
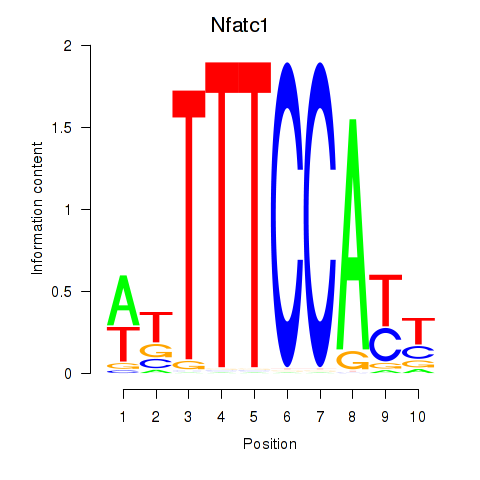
Transcription factors associated with Nfatc1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfatc1 | ENSMUSG00000033016.9 | Nfatc1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc1 | mm10_v2_chr18_-_80713062_80713080 | 0.23 | 4.8e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.4 | 46.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 6.5 | 25.9 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 5.9 | 17.7 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 4.2 | 12.6 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 3.0 | 5.9 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 2.5 | 7.6 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 2.4 | 31.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 2.2 | 9.0 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 2.2 | 13.3 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 2.2 | 6.6 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 2.1 | 10.5 | GO:1901526 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 2.0 | 6.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.9 | 5.6 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 1.8 | 12.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.7 | 13.5 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 1.6 | 4.9 | GO:1901228 | regulation of osteoclast proliferation(GO:0090289) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) negative regulation of bone development(GO:1903011) |
| 1.5 | 17.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 1.4 | 16.8 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 1.2 | 5.9 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 1.1 | 8.8 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 1.0 | 9.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.9 | 5.5 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.9 | 4.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.9 | 12.5 | GO:0072189 | ureter development(GO:0072189) |
| 0.9 | 6.2 | GO:0007320 | insemination(GO:0007320) |
| 0.9 | 2.6 | GO:0060084 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 0.9 | 2.6 | GO:0090135 | actin filament branching(GO:0090135) negative regulation of phospholipase C activity(GO:1900275) |
| 0.7 | 6.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.7 | 11.0 | GO:1902170 | cellular response to reactive nitrogen species(GO:1902170) |
| 0.7 | 5.5 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.6 | 3.7 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.6 | 28.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.6 | 3.6 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.6 | 2.4 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.6 | 0.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.5 | 1.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.4 | 7.4 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.4 | 2.5 | GO:0098734 | protein depalmitoylation(GO:0002084) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) macromolecule depalmitoylation(GO:0098734) |
| 0.3 | 3.7 | GO:0043586 | tongue development(GO:0043586) |
| 0.3 | 1.3 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.3 | 1.3 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.3 | 11.9 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.3 | 1.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 2.7 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 9.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 2.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 5.5 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 6.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.2 | 2.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.2 | 4.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 3339.9 | GO:0008150 | biological_process(GO:0008150) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 17.7 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 2.2 | 9.0 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 2.2 | 11.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 1.9 | 5.6 | GO:0071914 | prominosome(GO:0071914) |
| 1.0 | 4.1 | GO:0031673 | H zone(GO:0031673) |
| 1.0 | 21.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.9 | 12.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.9 | 8.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.8 | 11.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.7 | 9.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 7.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 3.6 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 2.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 1.9 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 11.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 3612.8 | GO:0005575 | cellular_component(GO:0005575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 4.4 | 17.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 2.7 | 24.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 2.7 | 10.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 2.1 | 10.5 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 2.0 | 7.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.5 | 6.0 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 1.4 | 5.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.3 | 9.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 1.3 | 7.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 1.2 | 3.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 1.0 | 12.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.9 | 3.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.7 | 25.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.7 | 41.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.6 | 5.8 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.6 | 5.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 4.9 | GO:0036122 | BMP binding(GO:0036122) |
| 0.5 | 11.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.4 | 7.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.4 | 13.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.3 | 1.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.3 | 13.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.3 | 4.0 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.3 | 5.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.3 | 1.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.3 | 2.6 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 7.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 4.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 2.6 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 1.4 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 2.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 4.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 1.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 3542.9 | GO:0003674 | molecular_function(GO:0003674) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 32.0 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.5 | 22.9 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.4 | 11.7 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 4.5 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 11.0 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 19.3 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.3 | 22.0 | PID_P73PATHWAY | p73 transcription factor network |
| 0.2 | 7.7 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 5.5 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 4.9 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 5.5 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.0 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.1 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.1 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 17.7 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.0 | 25.9 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.6 | 12.9 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.4 | 3.7 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.3 | 8.7 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.3 | 2.6 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 13.5 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.2 | 10.5 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 1.1 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 4.5 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 14.8 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 7.1 | REACTOME_RECRUITMENT_OF_MITOTIC_CENTROSOME_PROTEINS_AND_COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.1 | 3.6 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.3 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.7 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.3 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.7 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.9 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |