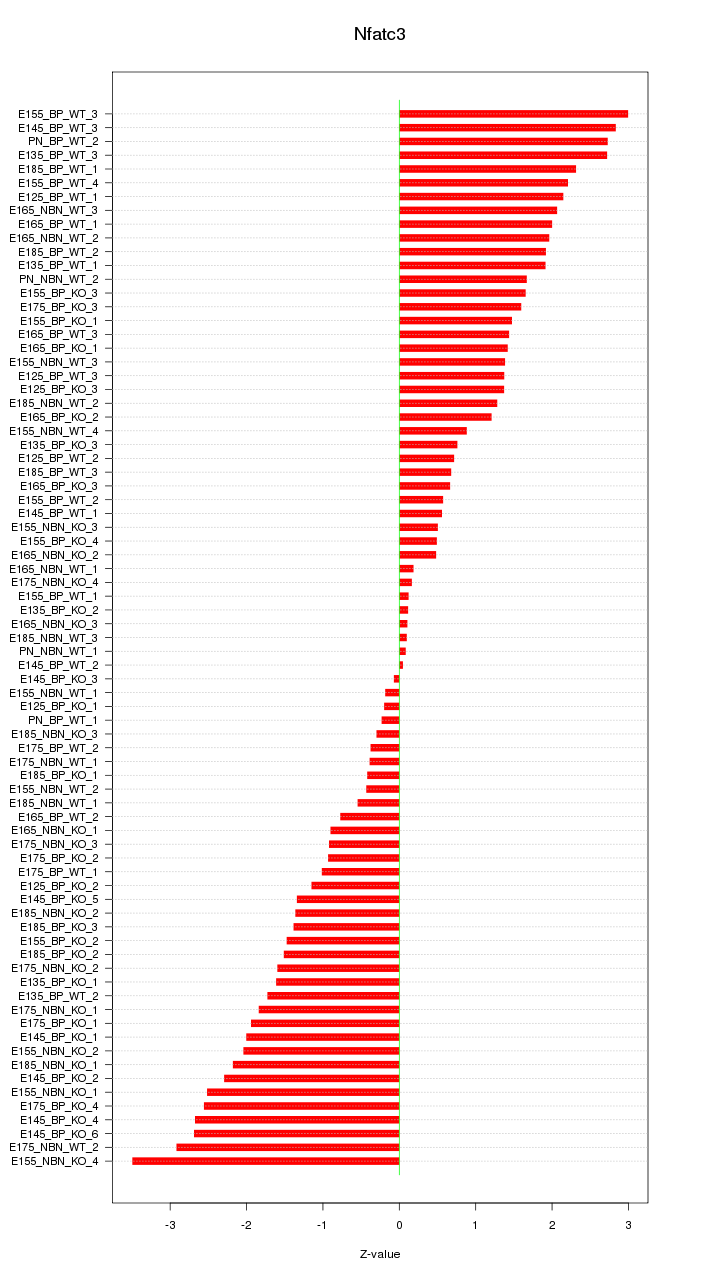
Motif ID: Nfatc3
Z-value: 1.578
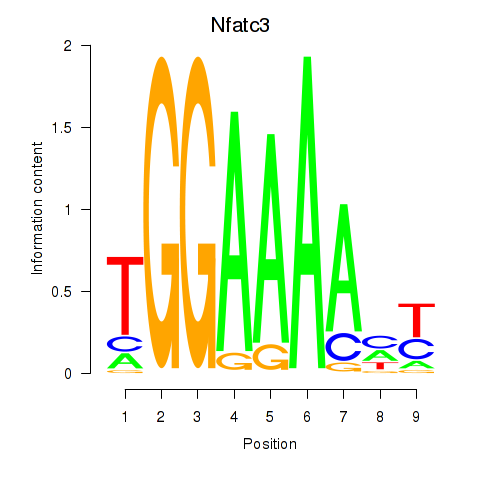
Transcription factors associated with Nfatc3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfatc3 | ENSMUSG00000031902.9 | Nfatc3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc3 | mm10_v2_chr8_+_106059562_106059623 | -0.02 | 8.7e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.6 | 19.8 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 3.8 | 26.7 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 3.7 | 11.1 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 2.5 | 7.4 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 2.2 | 13.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 2.2 | 10.8 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 2.2 | 8.6 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 2.0 | 10.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 1.7 | 5.2 | GO:0072554 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 1.4 | 5.6 | GO:2000319 | negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 1.3 | 1.3 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 1.2 | 10.8 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 1.2 | 9.5 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 1.1 | 3.4 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 1.1 | 6.4 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 1.0 | 6.3 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.9 | 2.7 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.9 | 4.4 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.9 | 2.6 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.8 | 2.4 | GO:1903903 | striated muscle atrophy(GO:0014891) positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of establishment of T cell polarity(GO:1903903) |
| 0.8 | 3.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.8 | 2.3 | GO:0003032 | detection of oxygen(GO:0003032) negative regulation of macrophage cytokine production(GO:0010936) |
| 0.8 | 0.8 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.8 | 6.8 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.7 | 6.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.7 | 3.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.7 | 2.0 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.6 | 1.9 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.6 | 1.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.6 | 1.9 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.6 | 4.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.6 | 1.8 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.6 | 2.9 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.6 | 2.3 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.5 | 3.7 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.5 | 10.7 | GO:0060579 | ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.5 | 3.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.5 | 4.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.5 | 3.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.5 | 2.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.5 | 5.0 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.5 | 1.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.5 | 2.8 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.5 | 2.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.5 | 3.2 | GO:0051461 | protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.4 | 1.8 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.4 | 2.2 | GO:0019659 | fermentation(GO:0006113) lactate biosynthetic process from pyruvate(GO:0019244) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.4 | 6.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 5.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 2.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 1.2 | GO:0009838 | abscission(GO:0009838) |
| 0.4 | 1.2 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.4 | 2.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.3 | 1.4 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 6.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.3 | 5.9 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.3 | 1.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.3 | 1.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.3 | 1.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.3 | 1.5 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.3 | 2.7 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.3 | 0.8 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.3 | 1.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.3 | 1.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.3 | 1.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.3 | 1.3 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.3 | 2.1 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.3 | 3.8 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.2 | 3.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 0.7 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.2 | 1.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 3.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.7 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.2 | 2.5 | GO:0043586 | tongue development(GO:0043586) |
| 0.2 | 0.7 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 1.1 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.2 | 5.4 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 1.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 0.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.2 | 0.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 1.2 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 1.8 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.2 | 2.7 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.2 | 0.6 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.2 | 1.5 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.2 | 0.7 | GO:0097298 | regulation of nucleus size(GO:0097298) |
| 0.2 | 4.0 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.2 | 2.7 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.2 | 3.9 | GO:0030575 | nuclear body organization(GO:0030575) |
| 0.2 | 0.7 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 1.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 0.8 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 | 1.4 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 2.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 1.6 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.1 | 1.8 | GO:0048754 | branching morphogenesis of an epithelial tube(GO:0048754) |
| 0.1 | 1.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 3.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 1.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 6.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.0 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 4.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.5 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.1 | 0.4 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.1 | 1.8 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 0.4 | GO:0010749 | regulation of nitric oxide mediated signal transduction(GO:0010749) |
| 0.1 | 1.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 2.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.9 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.1 | 0.6 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 1.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 4.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 4.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 3.5 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.1 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 3.0 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.8 | GO:0001823 | ureteric bud development(GO:0001657) mesonephros development(GO:0001823) mesonephric epithelium development(GO:0072163) mesonephric tubule development(GO:0072164) |
| 0.1 | 1.3 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 4.5 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.1 | 0.6 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.9 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 5.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 5.1 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.0 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.1 | 0.9 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 1.2 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.1 | 3.2 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 1.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.5 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.8 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 3.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 0.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.6 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.1 | 0.6 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 2.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 3.0 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 1.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:2001268 | keratinocyte migration(GO:0051546) regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) positive regulation of receptor binding(GO:1900122) negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 2.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 1.4 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 1.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.5 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.0 | 2.4 | GO:0050709 | negative regulation of protein secretion(GO:0050709) |
| 0.0 | 0.2 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.0 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.6 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 0.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 1.0 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.5 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.3 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.6 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 1.6 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.4 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 1.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.1 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 1.4 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.6 | GO:0016051 | carbohydrate biosynthetic process(GO:0016051) |
| 0.0 | 1.1 | GO:0000910 | cytokinesis(GO:0000910) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 1.0 | 5.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.8 | 5.9 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.8 | 3.2 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.8 | 3.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.7 | 2.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.6 | 2.8 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.5 | 5.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 2.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 3.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 2.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 2.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 6.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.3 | 6.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 5.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 1.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 1.2 | GO:1990131 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 2.7 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.2 | 3.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 14.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 2.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 3.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 5.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.2 | 0.6 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 2.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 11.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 8.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.4 | GO:0030934 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.1 | 1.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 2.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 5.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.0 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 3.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 3.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 3.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 2.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 3.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 0.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 3.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 5.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 6.5 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 3.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 3.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 3.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 4.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 7.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 12.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 0.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 1.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 8.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 4.2 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 9.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.1 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 2.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 6.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 3.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.8 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 75.2 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.1 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 2.9 | 8.6 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 2.5 | 7.4 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 2.4 | 9.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.6 | 6.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.1 | 3.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.0 | 2.9 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.9 | 2.8 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.9 | 4.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.8 | 10.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.8 | 10.0 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.6 | 3.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.6 | 1.9 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.6 | 2.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.6 | 1.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.6 | 4.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.6 | 2.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.6 | 2.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.6 | 1.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.5 | 2.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.5 | 5.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.5 | 1.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 2.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 3.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.4 | 1.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.4 | 3.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 2.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.4 | 1.6 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.4 | 1.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.4 | 1.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.4 | 2.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 1.0 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.3 | 1.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.3 | 1.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 1.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 19.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.3 | 6.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.3 | 5.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 1.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.3 | 1.0 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.3 | 3.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 7.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 3.0 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.2 | 0.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 0.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.2 | 2.7 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 2.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.9 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.2 | 3.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 5.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 1.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 1.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 2.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 4.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.2 | 1.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 7.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.2 | 3.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 0.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.5 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 2.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 2.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.6 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 4.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 2.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 1.3 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 2.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 4.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.0 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 1.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.6 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.8 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.5 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 2.7 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.1 | 1.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.8 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 11.1 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.1 | 1.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 2.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 2.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 9.2 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.1 | 0.9 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.1 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.8 | GO:0030346 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 6.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.2 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 1.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 2.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 8.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.8 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 1.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 2.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 8.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0001179 | RNA polymerase I transcription factor binding(GO:0001179) inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 1.2 | GO:0043028 | cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.7 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 1.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 16.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 4.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 1.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 2.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 5.7 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 0.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 34.5 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.4 | 9.8 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 6.3 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.3 | 4.3 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 15.3 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.2 | 10.0 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 4.5 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 15.9 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.2 | 6.3 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 4.4 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 1.5 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 4.1 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 6.3 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 4.8 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 5.1 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 1.8 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.1 | 2.0 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.2 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 4.5 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.9 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.6 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.8 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 0.9 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 5.5 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.1 | 3.3 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.8 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
| 0.1 | 3.3 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.1 | 2.5 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.9 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.1 | 6.4 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.3 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 4.8 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.1 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.2 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.9 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.0 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.7 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.5 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID_NFAT_3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 1.3 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.8 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.3 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | NABA_CORE_MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.2 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.6 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.7 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 10.8 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.5 | 1.9 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.5 | 7.4 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 2.1 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 5.2 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.3 | 4.1 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 9.3 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.2 | 2.3 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 5.9 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 1.8 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 6.9 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 6.3 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 3.9 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 11.6 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.4 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 4.0 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 11.6 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT_ATP_SYNTHESIS_BY_CHEMIOSMOTIC_COUPLING_AND_HEAT_PRODUCTION_BY_UNCOUPLING_PROTEINS_ | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 2.8 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 2.4 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.0 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.9 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 1.8 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.7 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.2 | REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILLING_BY_DNA_POL_IN_TC_NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 1.8 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 2.7 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.6 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.3 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.3 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 3.8 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.6 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_ACTIVATES_SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 1.2 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.3 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.8 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.2 | REACTOME_TRANSPORT_OF_MATURE_MRNA_DERIVED_FROM_AN_INTRONLESS_TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 0.9 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.6 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.8 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.6 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.1 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.3 | REACTOME_PROSTACYCLIN_SIGNALLING_THROUGH_PROSTACYCLIN_RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 7.1 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 3.0 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.3 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 3.5 | REACTOME_GASTRIN_CREB_SIGNALLING_PATHWAY_VIA_PKC_AND_MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.0 | 0.9 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.2 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.5 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.5 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 2.4 | REACTOME_SIGNALING_BY_EGFR_IN_CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.2 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.8 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.7 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME_PI_METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 0.3 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |