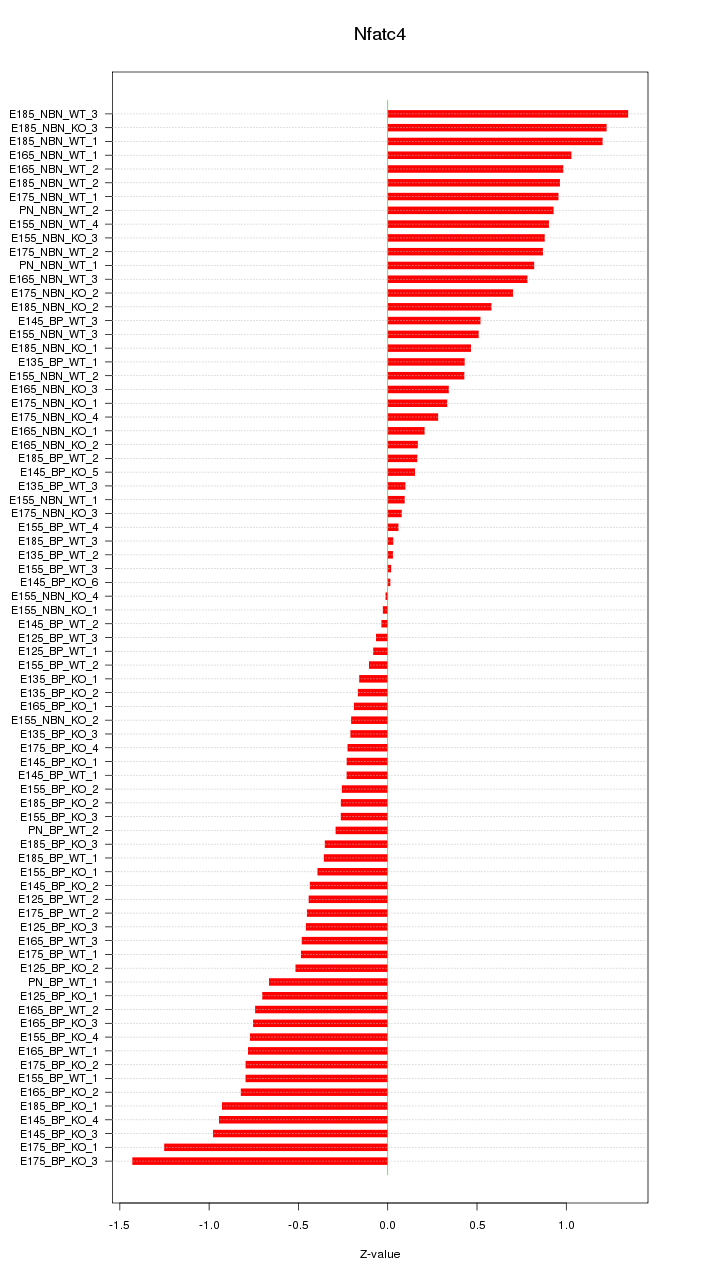
Motif ID: Nfatc4
Z-value: 0.620

Transcription factors associated with Nfatc4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfatc4 | ENSMUSG00000023411.5 | Nfatc4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc4 | mm10_v2_chr14_+_55824795_55824889 | -0.76 | 8.0e-16 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.3 | GO:0007521 | muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.4 | 4.1 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 1.3 | 3.9 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 1.2 | 3.7 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 1.0 | 16.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.7 | 4.3 | GO:1904587 | response to glycoprotein(GO:1904587) |
| 0.7 | 6.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.6 | 3.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.4 | 2.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.4 | 2.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.3 | 1.3 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.3 | 0.9 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.3 | 1.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 1.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.2 | 3.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 3.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 1.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.6 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.1 | 6.7 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 0.8 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 2.4 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.1 | 1.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 7.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.0 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 5.4 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.4 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 6.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 1.2 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 1.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 1.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 19.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 0.5 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 14.1 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 3.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 2.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 7.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 4.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 13.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.7 | 2.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.6 | 3.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.6 | 2.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 1.3 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 3.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.4 | 4.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 3.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 3.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 7.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 1.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 4.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 6.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 0.8 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 1.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.8 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 4.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.9 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 13.3 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.2 | 4.1 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.1 | 3.1 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 4.1 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.1 | 2.9 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 6.3 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 4.2 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.3 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.7 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.9 | PID_NOTCH_PATHWAY | Notch signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | REACTOME_HORMONE_LIGAND_BINDING_RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.4 | 12.9 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 3.7 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 7.1 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 3.7 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 5.5 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.1 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.7 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.9 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.0 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 5.3 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |