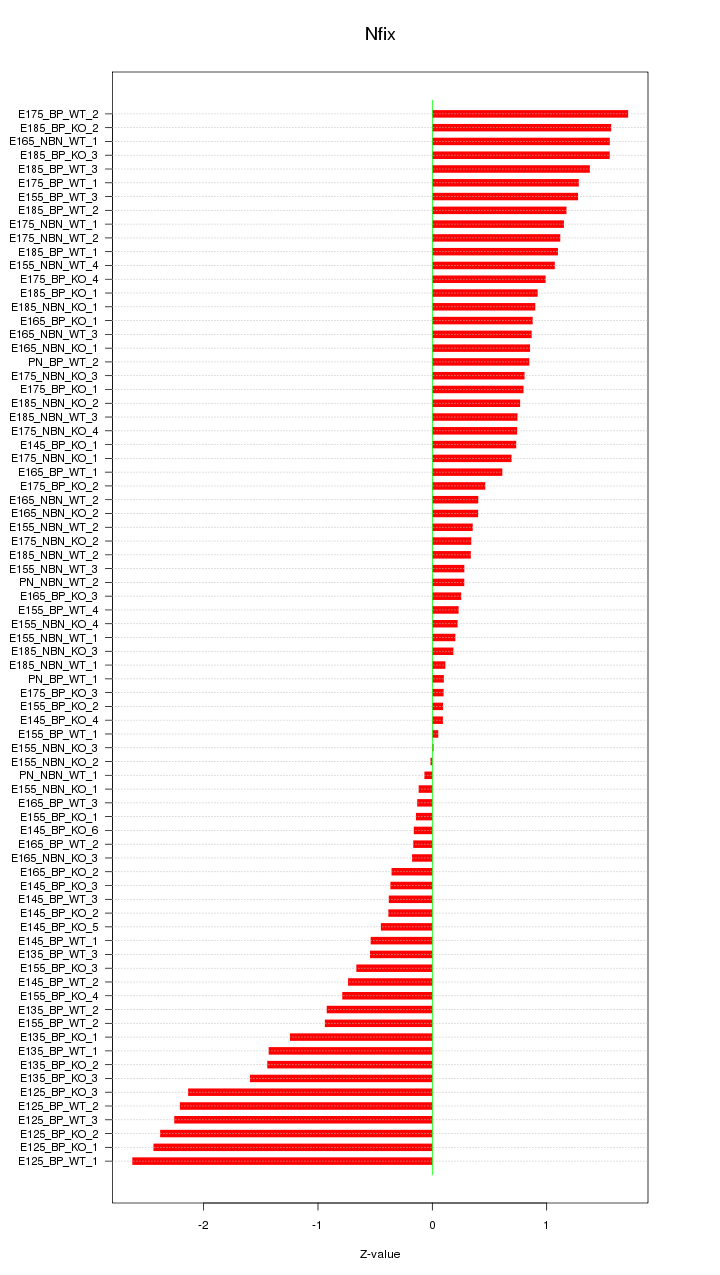
Motif ID: Nfix
Z-value: 1.013
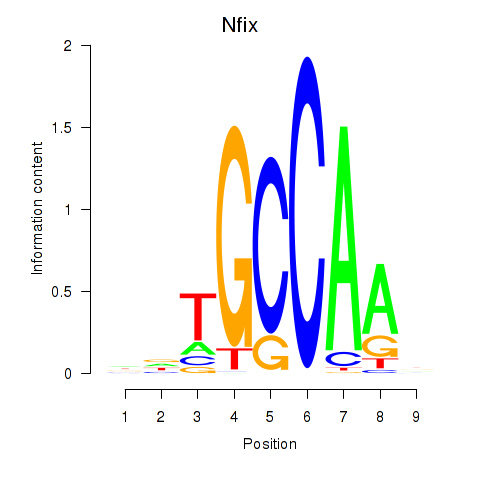
Transcription factors associated with Nfix:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfix | ENSMUSG00000001911.10 | Nfix |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfix | mm10_v2_chr8_-_84800024_84800283 | 0.91 | 2.7e-30 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.1 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 3.6 | 18.0 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 2.5 | 2.5 | GO:0003134 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) |
| 2.2 | 24.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 2.0 | 31.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.8 | 12.5 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 1.7 | 10.5 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 1.7 | 5.0 | GO:0007521 | muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.4 | 7.0 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 1.3 | 7.7 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 1.2 | 8.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.2 | 3.6 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 1.2 | 3.5 | GO:1905072 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 1.1 | 20.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 1.0 | 4.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.0 | 12.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 1.0 | 2.9 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.0 | 3.8 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 1.0 | 5.7 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.9 | 3.6 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.8 | 22.1 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.7 | 5.8 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.7 | 3.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.7 | 3.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.6 | 1.8 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.5 | 1.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.5 | 4.4 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.5 | 1.9 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.5 | 7.9 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.5 | 2.7 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.4 | 1.7 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.4 | 3.0 | GO:0098909 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.4 | 12.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.4 | 1.7 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.4 | 1.6 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.4 | 2.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.4 | 1.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.4 | 15.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.4 | 1.8 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 1.4 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.3 | 6.6 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.3 | 5.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.3 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 2.0 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 1.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 | 0.8 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.3 | 12.2 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.3 | 0.8 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 7.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 0.7 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) noradrenergic neuron differentiation(GO:0003357) |
| 0.2 | 2.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 1.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 2.0 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.2 | 4.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 0.6 | GO:0021998 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) regulation of cardiac ventricle development(GO:1904412) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.2 | 3.6 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 | 2.8 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.2 | 1.7 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.2 | 0.7 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.2 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 | 1.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 2.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.2 | 0.5 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.1 | 0.3 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.1 | 1.2 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.9 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 25.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 0.6 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.7 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 1.5 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 1.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.4 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.4 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 0.7 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 2.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.6 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.7 | GO:0060325 | head morphogenesis(GO:0060323) face morphogenesis(GO:0060325) |
| 0.1 | 2.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 1.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 4.8 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.1 | 1.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.6 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 1.7 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.1 | 2.2 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.5 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 1.0 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.9 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.7 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 1.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.7 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 1.9 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.2 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 2.4 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 1.5 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.7 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 1.9 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 1.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 1.5 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.6 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.6 | GO:0001890 | placenta development(GO:0001890) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 18.0 | GO:0043512 | inhibin A complex(GO:0043512) |
| 4.8 | 19.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 3.1 | 12.5 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 1.4 | 7.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.8 | 6.3 | GO:0097433 | dense body(GO:0097433) |
| 0.7 | 2.9 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.5 | 1.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 12.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.5 | 1.8 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.4 | 1.9 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 7.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 29.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.3 | 2.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 1.7 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.2 | 1.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.6 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.2 | 2.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 21.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 4.4 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 3.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 4.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.6 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 1.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 3.8 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.1 | 0.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.8 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 1.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.8 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 6.7 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 3.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 13.8 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 4.2 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 5.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 7.3 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.5 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 22.0 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 1.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 4.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.6 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 24.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 4.8 | 19.1 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 4.1 | 12.2 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 2.9 | 17.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.8 | 5.5 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 1.4 | 7.0 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 1.3 | 6.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.2 | 7.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.1 | 3.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.1 | 3.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 1.0 | 4.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.9 | 6.3 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.8 | 8.5 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.7 | 3.5 | GO:0005534 | galactose binding(GO:0005534) |
| 0.6 | 3.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.6 | 2.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.5 | 3.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.5 | 1.9 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.5 | 19.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.4 | 14.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.4 | 2.5 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.4 | 7.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.4 | 2.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.4 | 1.8 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.4 | 2.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.3 | 2.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 2.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.3 | 6.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.3 | 5.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 10.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 12.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 1.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 0.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.2 | 1.9 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 1.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 4.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.2 | 13.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 6.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 4.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 0.8 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.2 | 1.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 0.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.2 | 2.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 1.0 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 3.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 7.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.9 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 0.4 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.7 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 4.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 3.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.6 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.9 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 6.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 27.5 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 2.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 6.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 1.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 12.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 2.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 3.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 2.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 19.7 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.5 | 8.8 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 13.5 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.3 | 7.5 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.3 | 11.6 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.3 | 2.7 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.2 | 6.5 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 6.1 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 4.3 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.0 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 6.6 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.3 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 1.9 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.7 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.1 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.1 | 0.8 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.4 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.6 | PID_INTEGRIN1_PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.5 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.5 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID_PI3K_PLC_TRK_PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.0 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.7 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 18.0 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 3.1 | 24.6 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.7 | 10.8 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.5 | 7.0 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.5 | 4.1 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 7.9 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.3 | 12.8 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 11.8 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.3 | 6.6 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.3 | 2.1 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 6.1 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.3 | 2.6 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 15.2 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 1.9 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.2 | 1.6 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 5.3 | REACTOME_A_TETRASACCHARIDE_LINKER_SEQUENCE_IS_REQUIRED_FOR_GAG_SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.9 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.7 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 2.2 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.8 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 1.2 | REACTOME_NFKB_IS_ACTIVATED_AND_SIGNALS_SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 0.9 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 2.4 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 9.1 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 3.6 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.8 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 0.9 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.5 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.3 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.5 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.9 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 0.8 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.0 | 0.8 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 2.5 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.7 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.6 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.4 | REACTOME_SIGNALING_BY_EGFR_IN_CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.7 | REACTOME_SPHINGOLIPID_METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.3 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.0 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.2 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |