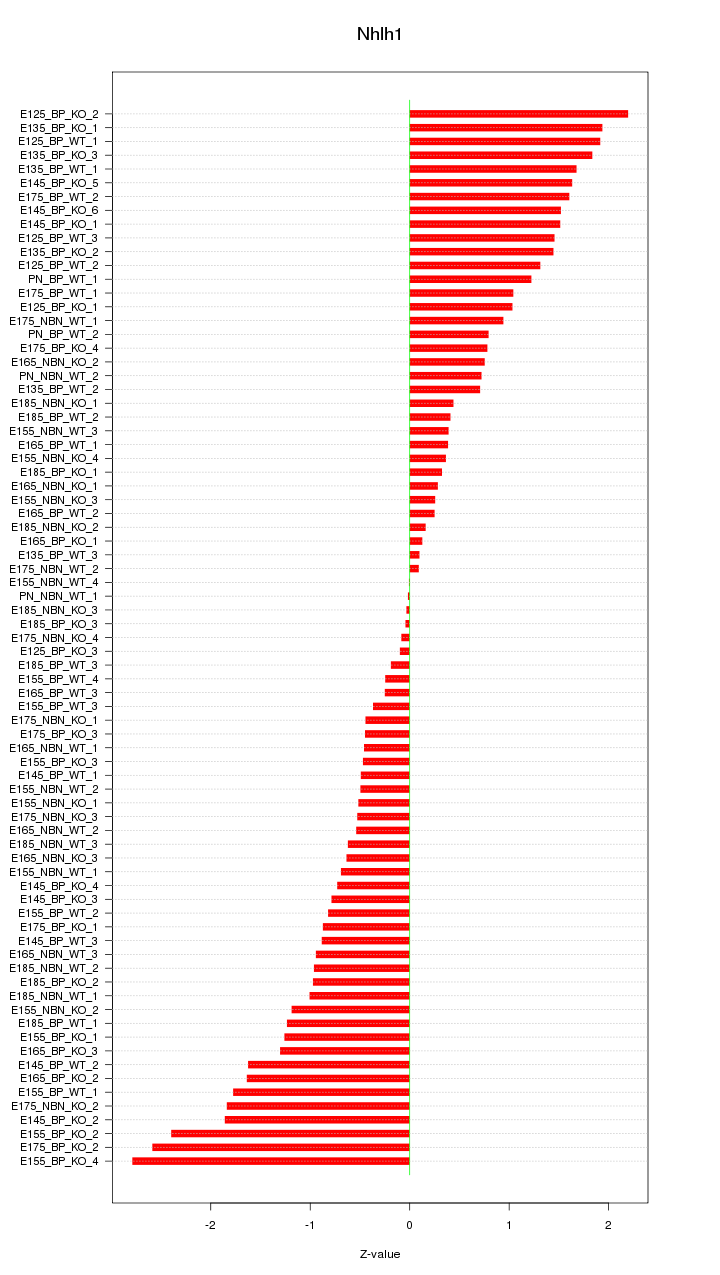
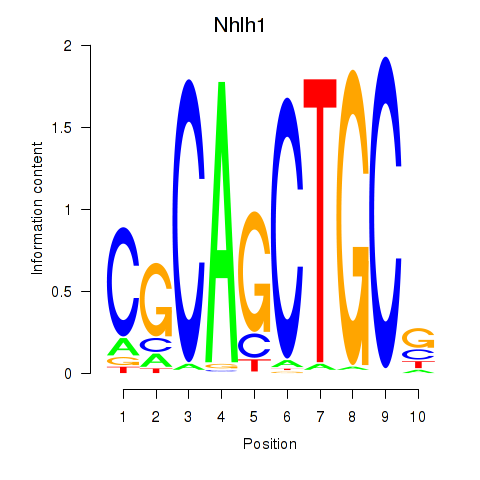
Motif ID: Nhlh1
Z-value: 1.113
Transcription factors associated with Nhlh1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nhlh1 | ENSMUSG00000051251.3 | Nhlh1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nhlh1 | mm10_v2_chr1_-_172057573_172057598 | 0.21 | 6.4e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.7 | 5.2 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 1.4 | 7.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.2 | 3.5 | GO:0070649 | meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 1.1 | 3.4 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.0 | 3.1 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 1.0 | 3.0 | GO:0072076 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 1.0 | 6.9 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 1.0 | 3.9 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.9 | 7.4 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.9 | 2.8 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.9 | 3.5 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.9 | 2.6 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.8 | 3.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.8 | 7.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.8 | 2.3 | GO:0072382 | minus-end-directed vesicle transport along microtubule(GO:0072382) |
| 0.8 | 0.8 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.8 | 3.8 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.7 | 6.0 | GO:0098712 | L-glutamate import across plasma membrane(GO:0098712) |
| 0.7 | 2.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.7 | 2.7 | GO:2000508 | regulation of dendritic cell chemotaxis(GO:2000508) |
| 0.7 | 2.0 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.7 | 3.9 | GO:0032439 | endosome localization(GO:0032439) |
| 0.7 | 3.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.6 | 3.2 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.6 | 5.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.6 | 3.8 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.6 | 1.9 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.6 | 5.4 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.6 | 3.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.6 | 1.8 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.6 | 0.6 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.6 | 4.1 | GO:0051461 | protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.6 | 2.9 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.6 | 2.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.6 | 4.5 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.6 | 6.7 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.5 | 2.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 0.5 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.5 | 1.0 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.5 | 1.0 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.5 | 1.9 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.5 | 3.3 | GO:0015791 | polyol transport(GO:0015791) |
| 0.5 | 0.9 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.5 | 4.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 2.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.4 | 1.8 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.4 | 0.9 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.4 | 2.6 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.4 | 1.3 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.4 | 1.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.4 | 2.9 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.4 | 5.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.4 | 1.2 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.4 | 4.6 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.4 | 3.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.4 | 2.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 0.8 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.4 | 1.6 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.4 | 1.2 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 0.4 | 2.0 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.4 | 2.0 | GO:0019230 | proprioception(GO:0019230) |
| 0.4 | 1.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.4 | 5.0 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.4 | 0.8 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 3.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.4 | 2.6 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.4 | 2.6 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.4 | 2.2 | GO:0060022 | hard palate development(GO:0060022) |
| 0.4 | 1.8 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.4 | 1.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.4 | 4.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.4 | 1.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.4 | 3.9 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.4 | 2.8 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.4 | 0.7 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 1.7 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.3 | 0.3 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.3 | 1.0 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.3 | 1.4 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.3 | 1.0 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.3 | 4.0 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.3 | 3.7 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) |
| 0.3 | 2.3 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 0.3 | 1.3 | GO:0060032 | notochord regression(GO:0060032) |
| 0.3 | 3.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 2.3 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.3 | 2.0 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.3 | 1.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.3 | 3.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 1.9 | GO:2000483 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.3 | 1.9 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 0.9 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K6-linked deubiquitination(GO:0044313) |
| 0.3 | 0.9 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.3 | 1.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.3 | 1.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.9 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.3 | 0.6 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.3 | 1.8 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.3 | 1.8 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.3 | 0.9 | GO:0046544 | regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) development of secondary male sexual characteristics(GO:0046544) |
| 0.3 | 1.2 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) regulation of potassium ion export(GO:1902302) |
| 0.3 | 0.6 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.3 | 0.3 | GO:0060295 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.3 | 1.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.3 | 0.9 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.3 | 1.4 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.3 | 0.6 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.3 | 3.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 1.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.3 | 0.8 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.3 | 0.8 | GO:0035984 | response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.3 | 1.1 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.3 | 12.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 1.9 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.3 | 6.5 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.3 | 0.5 | GO:0097709 | connective tissue replacement(GO:0097709) regulation of connective tissue replacement(GO:1905203) |
| 0.3 | 0.8 | GO:0045658 | eosinophil differentiation(GO:0030222) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.3 | 2.1 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.3 | 0.8 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.3 | 1.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.3 | 2.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.3 | 8.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.3 | 2.6 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.3 | 1.3 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.3 | 1.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.3 | 0.8 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 3.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.3 | 3.8 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 | 1.5 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 1.7 | GO:0098535 | positive regulation of centriole replication(GO:0046601) de novo centriole assembly(GO:0098535) |
| 0.2 | 1.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 0.7 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.2 | 0.7 | GO:0044782 | cilium organization(GO:0044782) |
| 0.2 | 2.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 0.2 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.2 | 0.9 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.2 | 2.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.5 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.2 | 0.7 | GO:0021649 | vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) sensory neuron axon guidance(GO:0097374) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.2 | 0.9 | GO:0048105 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.2 | 4.6 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.2 | 0.5 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 1.4 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.2 | 1.1 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.2 | 1.4 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.2 | 0.4 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.2 | 1.8 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.2 | 1.5 | GO:0046549 | phenol-containing compound catabolic process(GO:0019336) retinal cone cell development(GO:0046549) |
| 0.2 | 1.5 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 1.1 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.2 | 0.4 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.2 | 3.2 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.2 | 4.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 0.6 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.2 | 1.9 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.2 | 0.4 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 0.2 | 1.9 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 1.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 0.6 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.2 | 0.8 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.2 | 0.6 | GO:1903061 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.2 | 0.8 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.2 | 3.6 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.2 | 0.6 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 8.9 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.2 | 0.6 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.2 | 0.8 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) negative regulation of telomere capping(GO:1904354) |
| 0.2 | 1.2 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.2 | 1.5 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.2 | 1.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 2.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 0.9 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 0.8 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 2.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.2 | 1.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.2 | 1.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 0.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 0.7 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.2 | 0.5 | GO:0098885 | actin filament bundle distribution(GO:0070650) modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.2 | 1.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 0.3 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 | 1.0 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 1.9 | GO:0070922 | small RNA loading onto RISC(GO:0070922) |
| 0.2 | 0.2 | GO:0090594 | wound healing involved in inflammatory response(GO:0002246) inflammatory response to wounding(GO:0090594) |
| 0.2 | 1.9 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 1.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 1.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 1.0 | GO:0045629 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 0.8 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 0.5 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.2 | 0.3 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.2 | 1.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 0.3 | GO:0072053 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.2 | 1.6 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.2 | 0.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.2 | 2.9 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 0.2 | GO:0048819 | regulation of hair follicle maturation(GO:0048819) regulation of catagen(GO:0051794) |
| 0.2 | 5.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.4 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 0.7 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 | 0.7 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.9 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.1 | 0.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 2.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.6 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 | 1.3 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 2.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.8 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.0 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.1 | 0.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.5 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.1 | 0.8 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 0.8 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 0.4 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.6 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 1.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 1.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 1.7 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 1.0 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 1.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.2 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 | 1.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.3 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 0.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.5 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.5 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.5 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 1.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.3 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.1 | 1.0 | GO:1900194 | negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.4 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 2.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.2 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.7 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 1.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 2.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.4 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 2.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.8 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.1 | 0.3 | GO:0071415 | cellular response to purine-containing compound(GO:0071415) |
| 0.1 | 2.2 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.3 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of toll-like receptor 2 signaling pathway(GO:0034137) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 | 0.8 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.1 | 0.6 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 1.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.3 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 2.9 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.3 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 | 0.7 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.1 | 0.7 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 2.1 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 1.3 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 1.6 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 1.4 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 2.3 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 0.3 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.5 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.1 | 0.9 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 1.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 0.2 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.3 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 2.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.9 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.1 | 0.3 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.2 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.1 | 0.5 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.3 | GO:0046006 | positive regulation of activated T cell proliferation(GO:0042104) regulation of activated T cell proliferation(GO:0046006) |
| 0.1 | 0.3 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.3 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 | 0.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.9 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.1 | 1.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.4 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.1 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.8 | GO:0030388 | fructose 6-phosphate metabolic process(GO:0006002) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.6 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 1.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.4 | GO:0060482 | lobar bronchus development(GO:0060482) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 2.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.6 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.1 | 0.4 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 1.5 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of mitotic spindle localization(GO:0040001) |
| 0.1 | 1.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.2 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 1.3 | GO:0002011 | morphogenesis of an epithelial sheet(GO:0002011) |
| 0.1 | 0.7 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.9 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.4 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.1 | 0.7 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.9 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 1.0 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 8.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.6 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.5 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 1.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 1.4 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 0.5 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.4 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 0.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 2.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 0.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 1.3 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.8 | GO:0098868 | endochondral bone growth(GO:0003416) bone growth(GO:0098868) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.2 | GO:0035148 | tube formation(GO:0035148) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.6 | GO:0046856 | phospholipid dephosphorylation(GO:0046839) phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.5 | GO:0061245 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.0 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.7 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.1 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 0.1 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.0 | 0.1 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 0.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 1.1 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.3 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.8 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.5 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 1.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.2 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.1 | GO:2001053 | regulation of mesenchymal cell apoptotic process(GO:2001053) negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 | 0.6 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.0 | GO:0031960 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.0 | 0.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.3 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) regulation of metaphase/anaphase transition of cell cycle(GO:1902099) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 1.3 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 1.2 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.1 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.0 | 0.2 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.7 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.0 | 0.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.5 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.1 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.3 | GO:0002385 | organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.8 | GO:0051647 | nucleus localization(GO:0051647) |
| 0.0 | 0.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.3 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.9 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 1.0 | GO:1903051 | negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 1.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.5 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.1 | GO:0050812 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 1.8 | GO:0016331 | morphogenesis of embryonic epithelium(GO:0016331) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.3 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.3 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 1.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.2 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 1.1 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.2 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.3 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.3 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.3 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.3 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0001996 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 6.9 | GO:0032133 | interphase microtubule organizing center(GO:0031021) chromosome passenger complex(GO:0032133) |
| 1.0 | 4.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.9 | 6.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.9 | 2.6 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.7 | 3.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.6 | 1.9 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.6 | 5.9 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.6 | 1.7 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.5 | 3.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.5 | 3.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.5 | 4.5 | GO:0005883 | neurofilament(GO:0005883) |
| 0.5 | 2.9 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.5 | 2.5 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.4 | 1.3 | GO:0005940 | septin ring(GO:0005940) |
| 0.4 | 4.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 1.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.4 | 2.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 2.7 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.3 | 2.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 4.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 0.9 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.3 | 1.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.3 | 0.9 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.3 | 0.9 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.3 | 1.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 2.6 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 3.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 1.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.3 | 1.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 0.7 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.2 | 1.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 1.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 1.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 1.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 1.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.2 | 2.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 0.9 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.2 | 2.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 3.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 9.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 3.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 0.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 6.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 1.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 1.3 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 0.9 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 1.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.5 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 1.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 6.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 0.5 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.2 | 1.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 15.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.4 | GO:0000802 | transverse filament(GO:0000802) |
| 0.1 | 0.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.7 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 2.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 4.6 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 1.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 7.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 2.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.8 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 2.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.3 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.1 | 0.4 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 0.7 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.9 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.8 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.9 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.1 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.9 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.5 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 0.3 | GO:0097651 | phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 3.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.4 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 2.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.9 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 4.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 7.3 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 7.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 3.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 4.3 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 5.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.2 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 12.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 1.8 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 3.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 3.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 3.3 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 1.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) |
| 0.0 | 1.7 | GO:0035770 | ribonucleoprotein granule(GO:0035770) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.1 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 2.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 10.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 3.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.0 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 1.1 | 5.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.0 | 4.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.0 | 4.8 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.9 | 2.6 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.8 | 4.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.8 | 2.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.8 | 4.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.7 | 2.9 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.7 | 3.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.7 | 2.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.6 | 3.9 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.6 | 3.8 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.6 | 4.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.6 | 1.7 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.6 | 1.7 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.5 | 2.5 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.5 | 2.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.5 | 2.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.5 | 2.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.5 | 16.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 2.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.4 | 1.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.4 | 1.3 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.4 | 2.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.4 | 4.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 1.2 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.4 | 2.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.4 | 4.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.4 | 7.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.4 | 2.9 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.4 | 1.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.3 | 2.4 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 1.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 1.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.3 | 4.6 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.3 | 1.6 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.3 | 1.6 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.3 | 3.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 8.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.3 | 0.9 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.3 | 1.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 6.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.3 | 1.7 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 2.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 0.8 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.3 | 0.8 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.3 | 0.8 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.3 | 2.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.3 | 0.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.3 | 1.6 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.3 | 1.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 2.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 1.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.3 | 1.8 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 9.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 3.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 1.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 2.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 0.7 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.2 | 0.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 10.1 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 1.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 0.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 1.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.2 | 1.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 1.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 1.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.6 | GO:0051379 | epinephrine binding(GO:0051379) |
| 0.2 | 1.8 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 1.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 9.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 0.5 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.2 | 2.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 2.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 2.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 3.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 1.0 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.2 | 1.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.2 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 2.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 2.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 1.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 6.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 6.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 0.5 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 1.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.2 | 1.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.8 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.1 | 2.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 1.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0070976 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.1 | 2.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 4.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.8 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.1 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 1.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.8 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.8 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.0 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 2.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.6 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 2.0 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.7 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.5 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 1.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 2.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.3 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 2.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.1 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.1 | 0.3 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.1 | 0.9 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.0 | GO:0008227 | G-protein coupled amine receptor activity(GO:0008227) |
| 0.1 | 2.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.0 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.6 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.3 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 0.3 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 10.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 10.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.7 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 2.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 4.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.4 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.1 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.4 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 1.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 2.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 18.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.1 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 1.2 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.2 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 8.4 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) |
| 0.1 | 6.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 1.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.6 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 3.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 2.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 4.2 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 1.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 1.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 1.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 5.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 1.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.4 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 1.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0052866 | phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) uridylate kinase activity(GO:0009041) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 2.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0061135 | endopeptidase regulator activity(GO:0061135) |
| 0.0 | 0.2 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0015116 | sulfate transmembrane transporter activity(GO:0015116) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 10.7 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.5 | 1.1 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.5 | 2.1 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.4 | 16.9 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.3 | 12.4 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 3.0 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.2 | 11.1 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.2 | 1.5 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 3.1 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 0.9 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 16.9 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.2 | 0.5 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.6 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 5.0 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.1 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.1 | 0.4 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 3.8 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.1 | 4.4 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.5 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 4.2 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.7 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.8 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 1.9 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 0.4 | PID_INTEGRIN_CS_PATHWAY | Integrin family cell surface interactions |
| 0.1 | 3.5 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 2.1 | PID_PI3K_PLC_TRK_PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 3.3 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.1 | 0.9 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.7 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.0 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.7 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.3 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.5 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.3 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.1 | 1.6 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.5 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 3.2 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 0.5 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.5 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 2.2 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.0 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.0 | 1.7 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.3 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.4 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.5 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.1 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.2 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 0.7 | REACTOME_SIGNALING_BY_FGFR_IN_DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.5 | 2.2 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 3.1 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 1.7 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.3 | 1.6 | REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.3 | 2.1 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.3 | 4.2 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 7.3 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 0.2 | REACTOME_SIGNALING_BY_NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.2 | 7.1 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 5.0 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 5.0 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.2 | 4.8 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.2 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 8.3 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.2 | 2.7 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_LATE_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 3.5 | REACTOME_INCRETIN_SYNTHESIS_SECRETION_AND_INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.2 | 17.6 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 8.2 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 4.3 | REACTOME_TRAF6_MEDIATED_NFKB_ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.2 | 2.2 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 2.8 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 2.9 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.2 | 1.5 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 3.2 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 3.0 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 5.8 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.6 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.3 | REACTOME_CELL_SURFACE_INTERACTIONS_AT_THE_VASCULAR_WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 1.1 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.6 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.2 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.1 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 3.2 | REACTOME_DOUBLE_STRAND_BREAK_REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 1.7 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.1 | REACTOME_ABACAVIR_TRANSPORT_AND_METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.2 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 3.8 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 0.9 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.5 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.9 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.3 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.5 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.6 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.5 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 4.0 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.2 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.6 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.4 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.2 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 3.6 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 0.5 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.8 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.2 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 1.5 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 0.9 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.9 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.8 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.8 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 3.0 | REACTOME_SIGNALING_BY_NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.1 | 0.5 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.7 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.8 | REACTOME_CELL_CELL_JUNCTION_ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 0.5 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.7 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.2 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.8 | REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 2.3 | REACTOME_CLEAVAGE_OF_GROWING_TRANSCRIPT_IN_THE_TERMINATION_REGION_ | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.0 | 3.7 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.7 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.2 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.1 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.1 | REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME_SOS_MEDIATED_SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 4.2 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.5 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 1.5 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.4 | REACTOME_TRANSCRIPTIONAL_ACTIVITY_OF_SMAD2_SMAD3_SMAD4_HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 1.5 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.1 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.9 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 6.7 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.3 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_GLUCAGON_LIKE_PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.8 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.6 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.9 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |