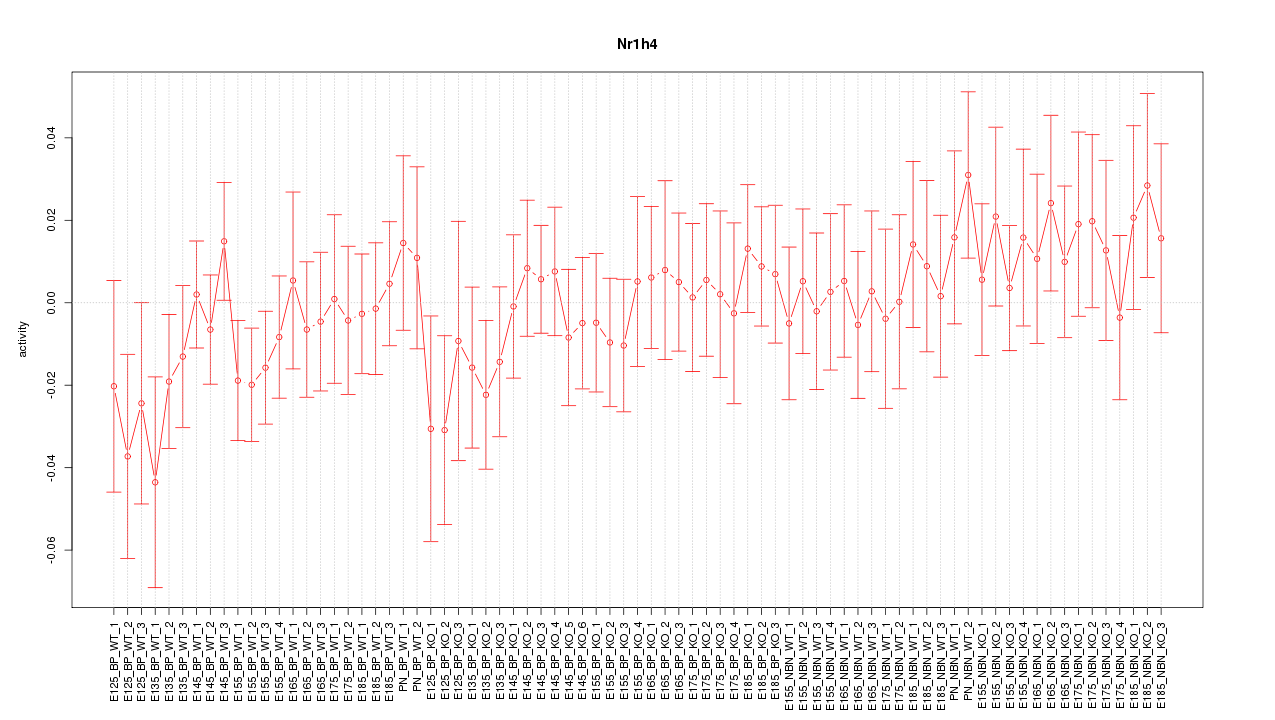
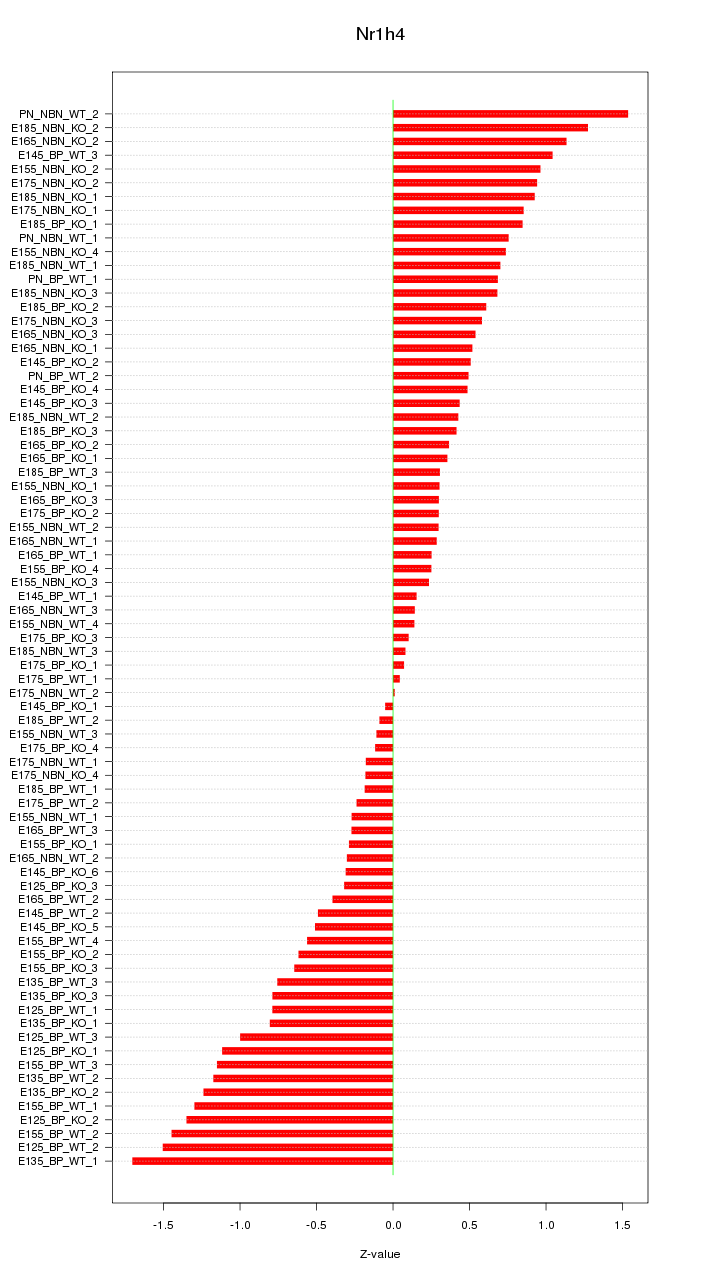
Motif ID: Nr1h4
Z-value: 0.710

Transcription factors associated with Nr1h4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr1h4 | ENSMUSG00000047638.9 | Nr1h4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.3 | 7.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.8 | 5.8 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.8 | 5.7 | GO:0010273 | detoxification of copper ion(GO:0010273) cellular response to zinc ion(GO:0071294) stress response to copper ion(GO:1990169) |
| 0.6 | 3.9 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.6 | 3.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.6 | 1.8 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.6 | 2.3 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.5 | 1.5 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.5 | 2.9 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.5 | 1.8 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.5 | 2.3 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.4 | 1.3 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.4 | 2.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.4 | 1.6 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.4 | 4.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 1.9 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 1.0 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.3 | 1.4 | GO:0051933 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.3 | 1.0 | GO:0001803 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.3 | 0.9 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.3 | 1.7 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.2 | 6.6 | GO:0030431 | sleep(GO:0030431) |
| 0.2 | 1.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 | 1.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 0.7 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.6 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.3 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 0.6 | GO:1903546 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.7 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.9 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 0.6 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 1.2 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 1.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 1.5 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.1 | 0.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 6.2 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 2.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 1.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.6 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.1 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 2.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.4 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.1 | 1.0 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 1.4 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.2 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 4.7 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 2.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 5.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 6.6 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.8 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.6 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 1.0 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.4 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.0 | 1.0 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.7 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 1.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0043512 | inhibin-betaglycan-ActRII complex(GO:0034673) inhibin A complex(GO:0043512) |
| 1.3 | 7.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.6 | 1.9 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.5 | 4.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 1.5 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.4 | 2.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.4 | 1.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.3 | 1.4 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.3 | 2.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.2 | 1.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 1.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.8 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 1.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 2.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 1.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.3 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 4.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.3 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.6 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 0.7 | GO:0030123 | AP-3 adaptor complex(GO:0030123) HOPS complex(GO:0030897) |
| 0.1 | 4.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.6 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 1.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 8.1 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 7.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 3.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.6 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.6 | GO:0043197 | dendritic spine(GO:0043197) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.9 | 8.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.9 | 7.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.8 | 3.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.6 | 4.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.5 | 4.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 2.5 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.5 | 1.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.5 | 1.8 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.4 | 3.9 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.4 | 2.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.4 | 1.5 | GO:0004096 | catalase activity(GO:0004096) |
| 0.4 | 1.5 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 4.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 2.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 1.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 0.9 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.2 | 0.9 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.2 | 1.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 4.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 5.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 5.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.3 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.1 | 1.1 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 1.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 1.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 1.0 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 2.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 1.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 2.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 5.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 2.7 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 9.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 7.8 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.9 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 4.2 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 5.0 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.6 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.9 | 8.9 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.6 | 4.4 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 14.5 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.3 | 3.2 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 2.9 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 2.5 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.3 | REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 6.6 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 4.7 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.8 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.5 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 2.3 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.5 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.9 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.6 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.7 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 1.2 | REACTOME_ELONGATION_ARREST_AND_RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.4 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.4 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |