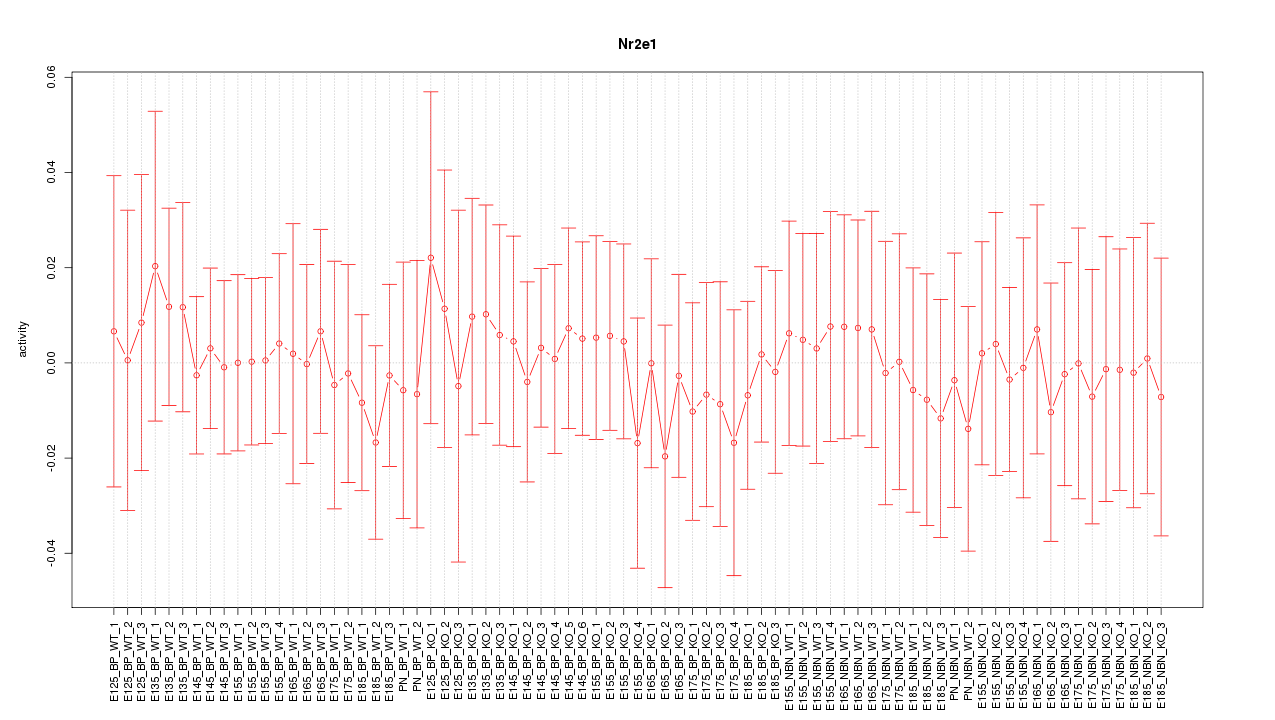
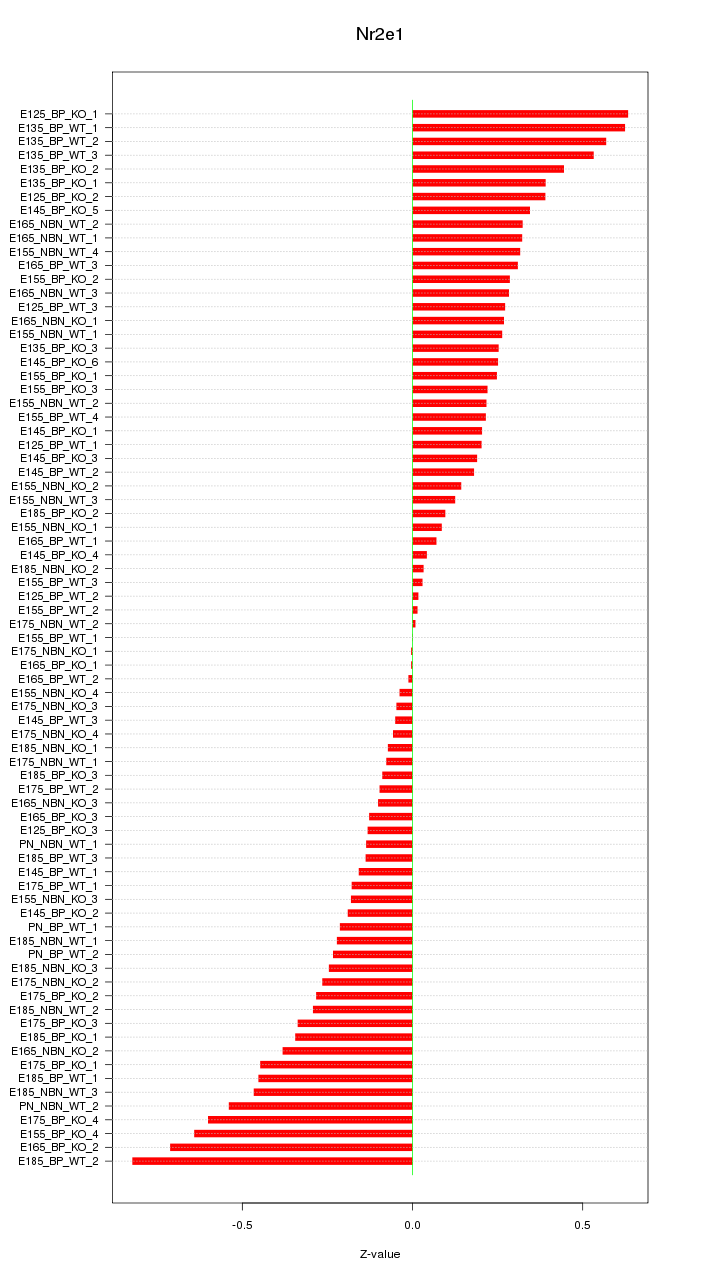
Motif ID: Nr2e1
Z-value: 0.308

Transcription factors associated with Nr2e1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr2e1 | ENSMUSG00000019803.5 | Nr2e1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2e1 | mm10_v2_chr10_-_42583628_42583639 | 0.11 | 3.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0031622 | positive regulation of fever generation(GO:0031622) trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.4 | 2.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.2 | 2.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 1.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 2.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.3 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 1.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.4 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 1.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 1.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 1.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.2 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 2.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 1.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 1.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 1.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 2.2 | GO:0008144 | drug binding(GO:0008144) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.4 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.2 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 1.0 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME_ACTIVATION_OF_IRF3_IRF7_MEDIATED_BY_TBK1_IKK_EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 2.2 | REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.3 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |