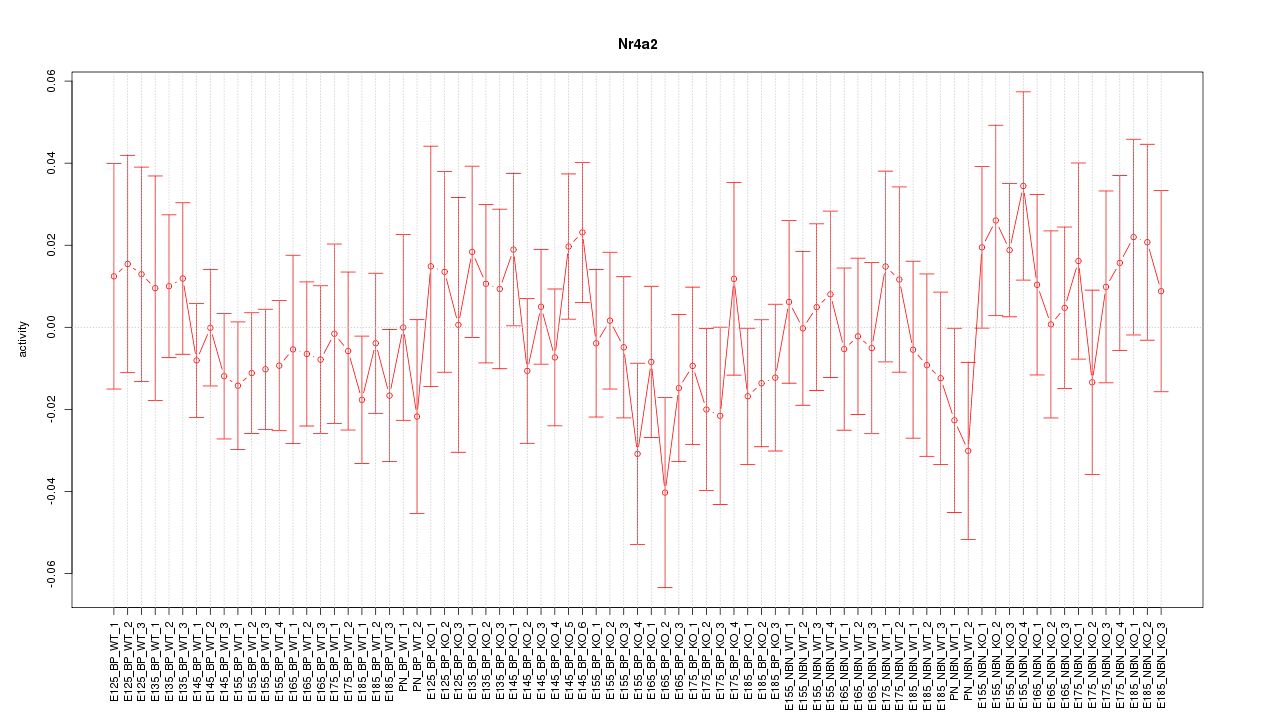
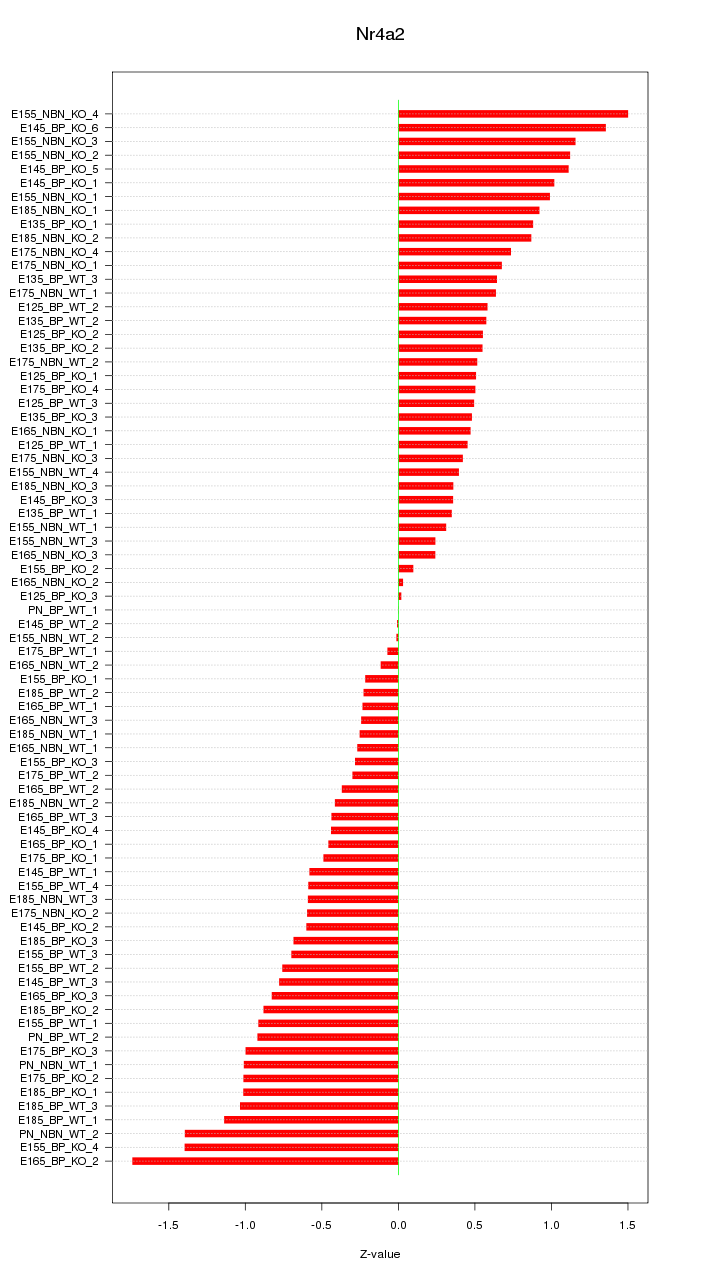
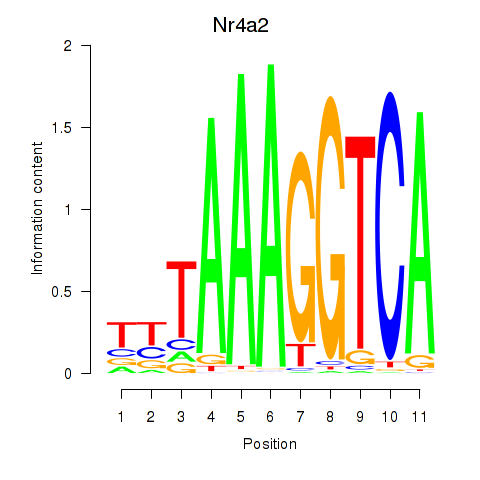
Motif ID: Nr4a2
Z-value: 0.722
Transcription factors associated with Nr4a2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nr4a2 | ENSMUSG00000026826.7 | Nr4a2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr4a2 | mm10_v2_chr2_-_57124003_57124071 | 0.70 | 9.0e-13 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 1.9 | 5.8 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.7 | 3.7 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.7 | 2.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.6 | 5.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.6 | 1.7 | GO:0071336 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.5 | 4.9 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.5 | 1.6 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.5 | 2.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.5 | 1.5 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.5 | 7.5 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.4 | 1.8 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.4 | 4.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.4 | 1.2 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) negative regulation of protein import into nucleus, translocation(GO:0033159) regulation of oligodendrocyte apoptotic process(GO:1900141) negative regulation of oligodendrocyte apoptotic process(GO:1900142) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.4 | 3.6 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.3 | 3.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 0.6 | GO:0015866 | ADP transport(GO:0015866) |
| 0.2 | 2.7 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 1.8 | GO:0032306 | regulation of prostaglandin secretion(GO:0032306) |
| 0.2 | 1.7 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.2 | 3.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.2 | 1.4 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.2 | 1.8 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.7 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 4.8 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 0.3 | GO:0070973 | COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.5 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 4.1 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 2.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 8.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.9 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 3.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.6 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.6 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 3.8 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.1 | 0.4 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 1.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.7 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 1.7 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.0 | 0.6 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.2 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 1.4 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.5 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.0 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.4 | 3.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 5.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 1.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 1.8 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 1.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 7.5 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 5.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 1.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.3 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 7.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 4.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.5 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 4.8 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 5.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.3 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0005930 | axoneme(GO:0005930) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.7 | 3.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.7 | 7.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.7 | 8.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.6 | 3.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.5 | 5.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.5 | 2.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.5 | 2.0 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.5 | 3.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.4 | 1.7 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 1.6 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 3.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.7 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 7.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.5 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.7 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.1 | 1.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 2.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.6 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 4.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 5.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 2.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 3.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 2.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 3.4 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.8 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 4.2 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.8 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.1 | 3.8 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.8 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.7 | PID_PI3K_PLC_TRK_PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 2.1 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.0 | 5.9 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 2.4 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.1 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.2 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.5 | 1.4 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 5.8 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 4.1 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 3.3 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 2.5 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.1 | 3.3 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.7 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 2.1 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 7.4 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.2 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.4 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.5 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.8 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 6.3 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME_METABOLISM_OF_STEROID_HORMONES_AND_VITAMINS_A_AND_D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.3 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.6 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.2 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.9 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.6 | REACTOME_P75_NTR_RECEPTOR_MEDIATED_SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |