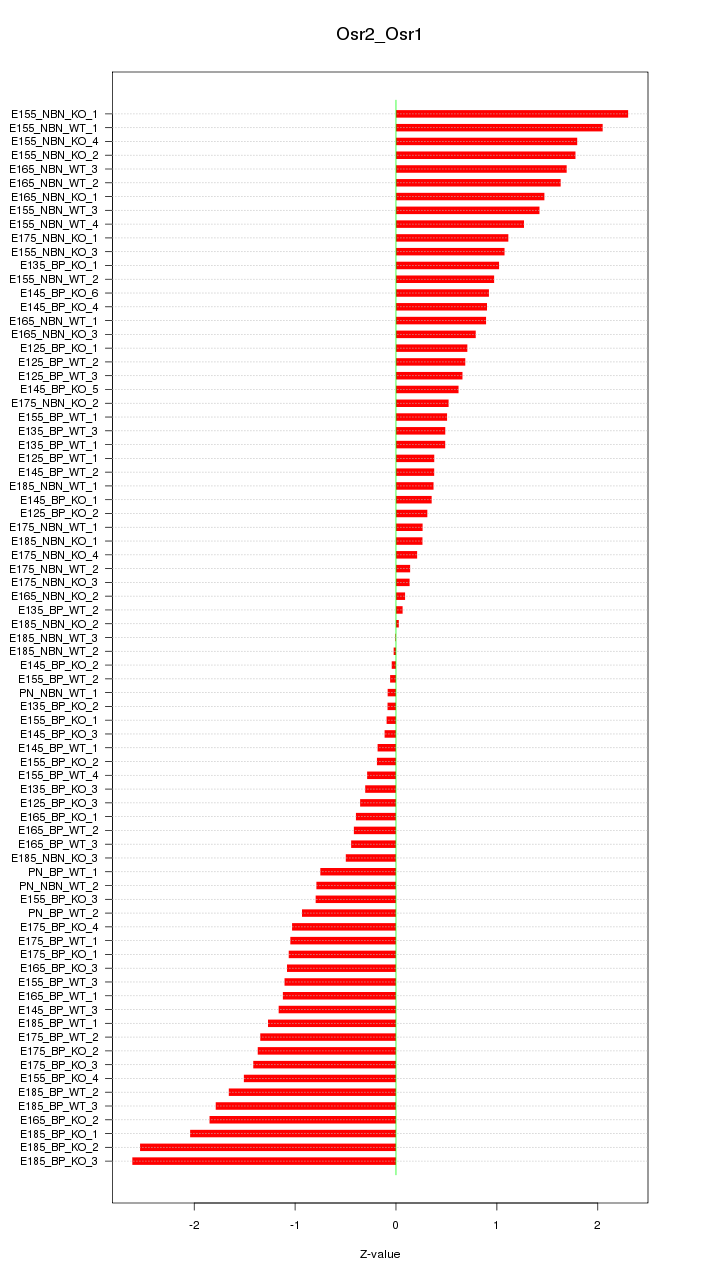
Motif ID: Osr2_Osr1
Z-value: 1.062
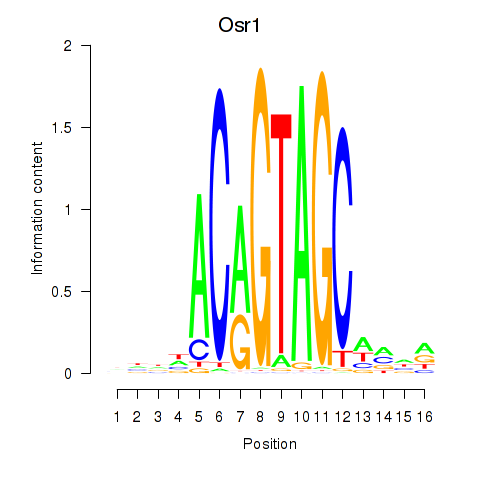
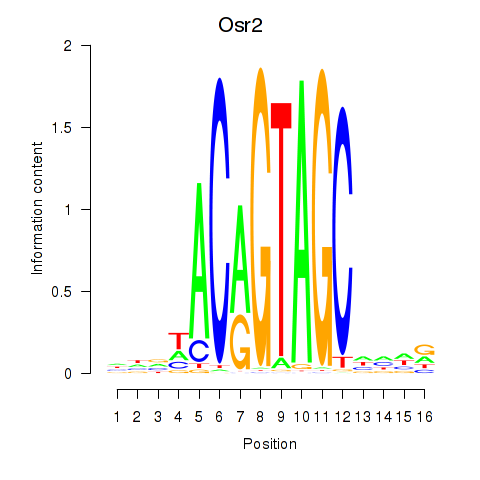
Transcription factors associated with Osr2_Osr1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Osr1 | ENSMUSG00000048387.7 | Osr1 |
| Osr2 | ENSMUSG00000022330.4 | Osr2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Osr1 | mm10_v2_chr12_+_9574437_9574448 | 0.19 | 1.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 20.3 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 1.8 | 7.2 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 1.1 | 3.2 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.9 | 4.7 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.9 | 4.5 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.8 | 7.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.7 | 6.5 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.7 | 12.3 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.7 | 2.0 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.5 | 2.2 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.5 | 1.6 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.5 | 1.5 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.5 | 2.5 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.5 | 2.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.5 | 1.0 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.5 | 6.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.5 | 3.2 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.4 | 1.8 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.4 | 1.5 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 | 1.4 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.4 | 2.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.3 | 1.7 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.3 | 1.0 | GO:0006550 | isoleucine catabolic process(GO:0006550) acetyl-CoA catabolic process(GO:0046356) ketone body catabolic process(GO:0046952) |
| 0.3 | 11.5 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.3 | 1.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 0.9 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.3 | 1.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 0.7 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.2 | 1.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.2 | 5.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 0.6 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.2 | 3.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.2 | 1.8 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.2 | 0.5 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.2 | 1.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.9 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.1 | 4.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 7.8 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.7 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.6 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 1.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.3 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.4 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.6 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 1.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.9 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.1 | 0.6 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.6 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.1 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.5 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.1 | GO:0033864 | positive regulation of NAD(P)H oxidase activity(GO:0033864) |
| 0.1 | 4.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 1.7 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 1.0 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 1.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 1.0 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 1.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.1 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.3 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.3 | GO:1903894 | regulation of IRE1-mediated unfolded protein response(GO:1903894) |
| 0.0 | 0.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 2.2 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.4 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 4.8 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.5 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 1.1 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 1.7 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.5 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.7 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.7 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 1.8 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.7 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.8 | 3.4 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.6 | 1.9 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.5 | 2.5 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.4 | 1.7 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.4 | 1.5 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.3 | 5.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 4.0 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.3 | 2.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 1.8 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 6.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 0.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 2.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 5.6 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 2.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 1.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 2.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.2 | 15.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 2.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 10.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 16.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 3.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 4.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 15.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 5.1 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 7.9 | GO:0097458 | neuron part(GO:0097458) |
| 0.0 | 4.1 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) C-X-C chemokine binding(GO:0019958) |
| 1.8 | 12.3 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.3 | 4.0 | GO:0034437 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 0.7 | 2.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.7 | 2.0 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.5 | 1.6 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.5 | 3.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.5 | 1.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.4 | 1.8 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.4 | 7.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.3 | 0.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.3 | 0.9 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 5.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 2.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.2 | 3.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 6.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 3.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 1.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 0.6 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 1.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 1.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 1.6 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.2 | 1.0 | GO:0016453 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.1 | 4.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 2.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 6.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.7 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 1.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 2.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.5 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 2.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 3.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 4.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 18.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.8 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 4.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.7 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 1.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 2.1 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 1.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.0 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 2.8 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 2.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 17.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.1 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 4.0 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.4 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.1 | 4.5 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 4.4 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.4 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.2 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.5 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 1.5 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.0 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.0 | 5.2 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.8 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 1.5 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 4.0 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.0 | 0.8 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.8 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.0 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.1 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.3 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.3 | 5.2 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 1.8 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.3 | 1.0 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 6.1 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.0 | REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.8 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.1 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.4 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.7 | REACTOME_SIGNALLING_TO_P38_VIA_RIT_AND_RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 14.2 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.9 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 4.7 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.5 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.8 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 0.6 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.4 | REACTOME_PLC_BETA_MEDIATED_EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 1.0 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.9 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.7 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 2.0 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.6 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 7.0 | REACTOME_METABOLISM_OF_CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.7 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.5 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.9 | REACTOME_INWARDLY_RECTIFYING_K_CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.9 | REACTOME_PHASE_II_CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 1.2 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.2 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |