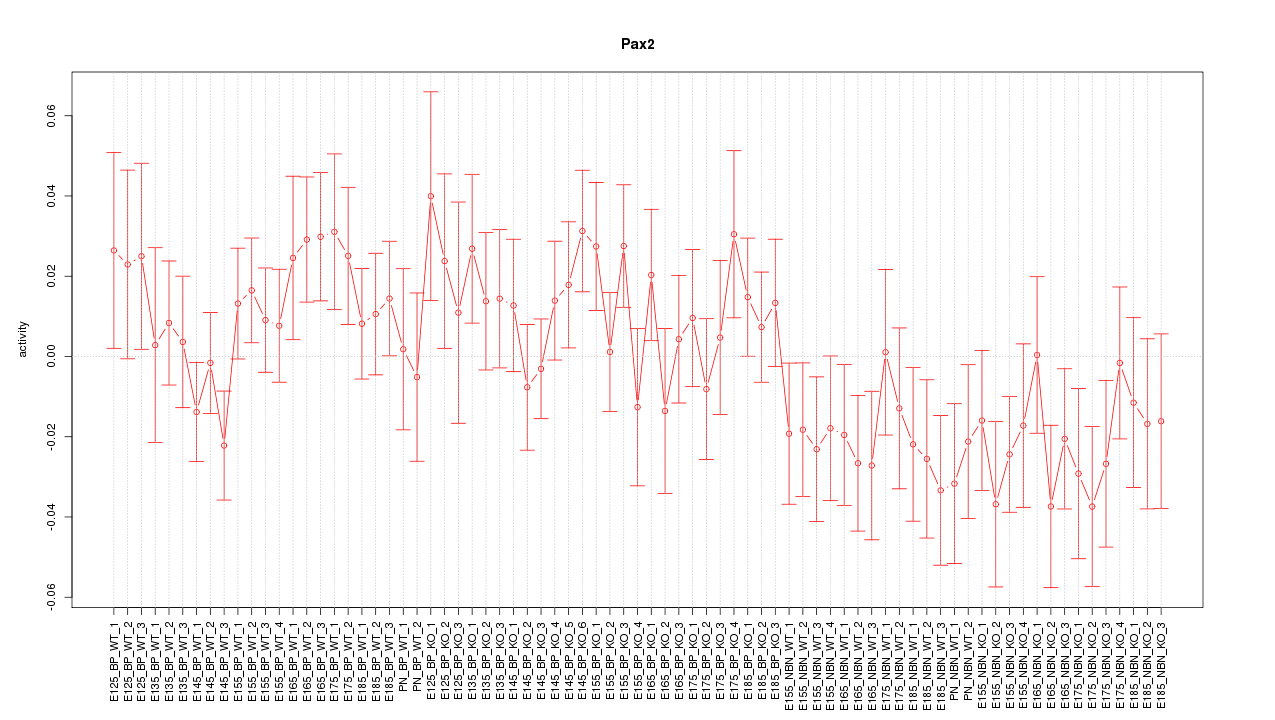
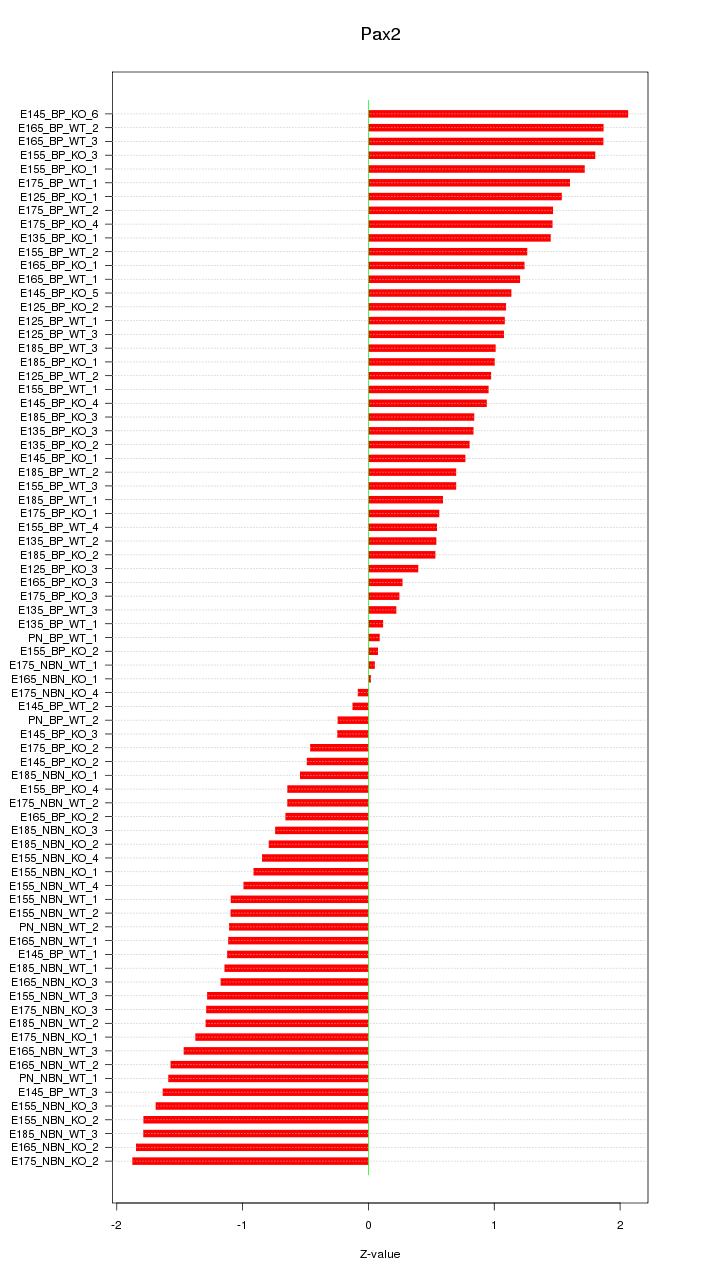
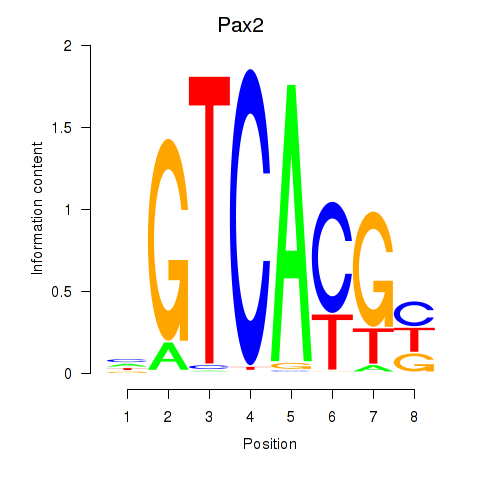
Motif ID: Pax2
Z-value: 1.117
Transcription factors associated with Pax2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pax2 | ENSMUSG00000004231.9 | Pax2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 17.8 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 3.3 | 9.8 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 2.6 | 15.7 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 2.6 | 12.9 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 2.4 | 9.6 | GO:0010288 | response to lead ion(GO:0010288) |
| 1.6 | 4.8 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 1.5 | 6.0 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 1.4 | 4.1 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 1.3 | 6.7 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 1.3 | 4.0 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 1.2 | 3.7 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 1.2 | 6.1 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) negative regulation of protein kinase C signaling(GO:0090038) |
| 1.2 | 4.8 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.2 | 3.6 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 1.1 | 7.9 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.1 | 3.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 1.0 | 6.9 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 1.0 | 4.8 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.9 | 2.6 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.9 | 2.6 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.8 | 4.0 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.8 | 4.8 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.7 | 11.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.6 | 2.5 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.6 | 4.7 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.5 | 1.5 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.5 | 4.4 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.5 | 2.8 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 1.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.4 | 1.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.4 | 1.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.4 | 6.3 | GO:0060746 | parental behavior(GO:0060746) |
| 0.4 | 3.3 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.4 | 1.8 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.3 | 4.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.3 | 1.0 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.3 | 1.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.3 | 1.8 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 | 4.5 | GO:1904293 | negative regulation of ERAD pathway(GO:1904293) |
| 0.3 | 1.8 | GO:0035878 | nail development(GO:0035878) |
| 0.3 | 2.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.3 | 2.8 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.3 | 2.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 1.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.3 | 1.8 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 | 0.9 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 9.5 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.2 | 2.3 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.2 | 12.7 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.2 | 0.6 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.2 | 2.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.9 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.2 | 2.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 4.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.2 | 8.0 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.2 | 3.9 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.2 | 5.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 2.7 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.2 | 2.3 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.2 | 2.8 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 2.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 2.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 3.3 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 2.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 4.6 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.1 | 2.1 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 1.7 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 3.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 5.0 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 4.8 | GO:0060351 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.1 | 0.6 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 1.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 2.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 1.5 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 6.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.0 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 5.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.9 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 3.5 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 2.0 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.1 | 2.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.2 | GO:1904395 | synaptic growth at neuromuscular junction(GO:0051124) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.1 | 0.4 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.1 | 1.9 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 4.9 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.8 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 7.0 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.1 | 0.5 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.9 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 7.4 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 2.9 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 1.7 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 1.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 2.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 3.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 2.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 3.1 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 2.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 3.6 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 3.7 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.9 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.6 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) fear response(GO:0042596) |
| 0.0 | 0.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 2.4 | 9.6 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 1.2 | 15.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) mitotic spindle midzone(GO:1990023) |
| 1.0 | 4.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.9 | 3.6 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.7 | 2.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.7 | 3.6 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.6 | 1.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.6 | 6.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.6 | 6.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.5 | 3.6 | GO:0001740 | Barr body(GO:0001740) |
| 0.4 | 4.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.4 | 3.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.4 | 1.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 1.8 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.3 | 11.9 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.3 | 3.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 2.0 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.3 | 1.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 3.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 1.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 4.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 2.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 1.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.2 | 1.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 3.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 1.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 4.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.4 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 4.9 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 10.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 4.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 4.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 2.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.3 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.1 | 7.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 5.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 3.0 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 3.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 3.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 4.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 4.0 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.7 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 0.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 5.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.0 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 2.5 | GO:0005813 | centrosome(GO:0005813) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.9 | 17.8 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 3.1 | 15.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.2 | 4.8 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.1 | 11.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.8 | 4.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.8 | 3.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.8 | 3.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.8 | 6.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.7 | 2.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.6 | 2.5 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.6 | 1.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.6 | 2.8 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.5 | 2.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 7.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 2.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.5 | 2.8 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.4 | 4.9 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 1.3 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.4 | 1.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.4 | 2.4 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.4 | 6.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.4 | 3.6 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.4 | 1.8 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 2.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.3 | 2.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.3 | 7.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 1.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 1.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 4.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 5.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 6.8 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 9.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 4.1 | GO:0008432 | JUN kinase binding(GO:0008432) co-SMAD binding(GO:0070410) |
| 0.2 | 1.8 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 7.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 0.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 0.5 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.2 | 0.6 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.2 | 0.9 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 1.6 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 5.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 3.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 3.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 6.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.4 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 3.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 3.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.8 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.9 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 36.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 1.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 14.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 4.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 2.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 3.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 8.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 3.0 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 41.4 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 1.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.0 | GO:0019905 | syntaxin binding(GO:0019905) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.6 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 11.0 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.4 | 4.2 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 18.9 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.2 | 1.8 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 7.9 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.1 | 2.1 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.6 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.1 | 3.5 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.1 | 3.5 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 4.0 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 3.5 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.9 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.0 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.1 | 4.1 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.1 | 2.3 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.4 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 4.5 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.8 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.3 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 2.0 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.1 | 3.6 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 2.3 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 2.0 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | REACTOME_ARMS_MEDIATED_ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.8 | 7.9 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.6 | 3.5 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.5 | 11.0 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.4 | 6.1 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.4 | 4.0 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 20.6 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 12.9 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.3 | 2.8 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 1.9 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 8.7 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 5.7 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 5.7 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 2.4 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.9 | REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 8.2 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 4.1 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 3.5 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.3 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 8.0 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 2.0 | REACTOME_TRANSPORT_OF_MATURE_MRNA_DERIVED_FROM_AN_INTRONLESS_TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 8.0 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 5.0 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.3 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.9 | REACTOME_VIF_MEDIATED_DEGRADATION_OF_APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 2.2 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 2.3 | REACTOME_SIGNALING_BY_ILS | Genes involved in Signaling by Interleukins |
| 0.0 | 1.8 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |