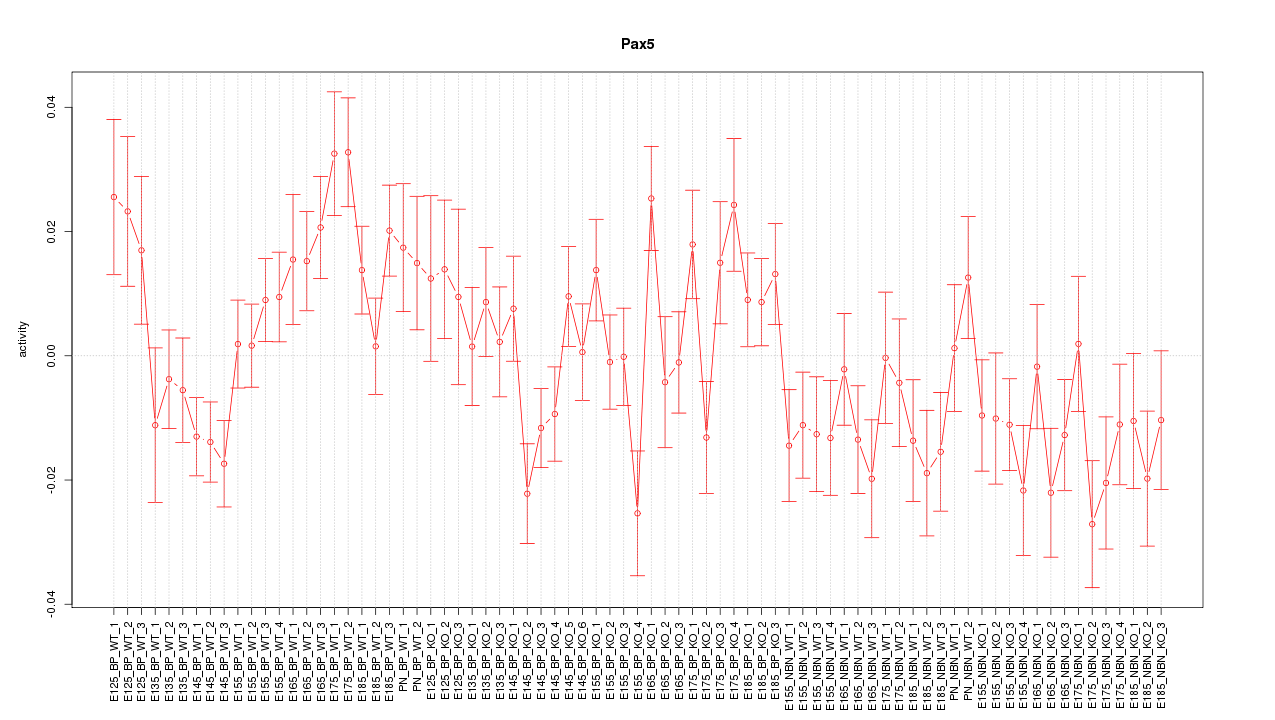
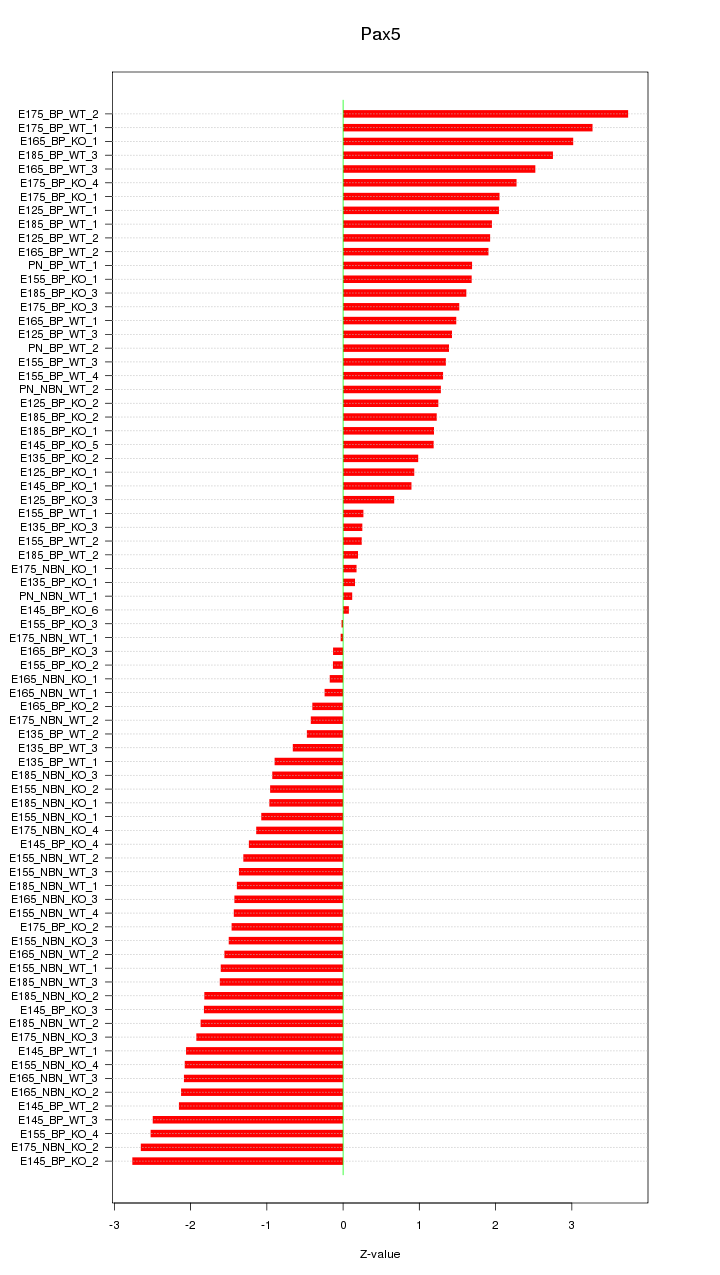
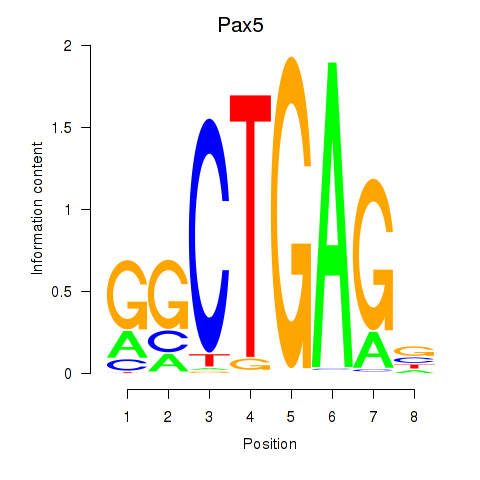
Motif ID: Pax5
Z-value: 1.600
Transcription factors associated with Pax5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pax5 | ENSMUSG00000014030.9 | Pax5 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax5 | mm10_v2_chr4_-_44710408_44710418 | 0.28 | 1.2e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.5 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 4.8 | 14.4 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 4.0 | 8.1 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 3.6 | 14.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 3.2 | 9.5 | GO:0071579 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) regulation of zinc ion transport(GO:0071579) |
| 3.0 | 3.0 | GO:0060067 | cervix development(GO:0060067) |
| 2.5 | 7.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 2.3 | 11.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 2.1 | 4.3 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 2.1 | 23.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 1.8 | 5.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.8 | 8.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 1.7 | 17.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 1.6 | 4.7 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 1.5 | 7.7 | GO:0042891 | antibiotic transport(GO:0042891) |
| 1.5 | 1.5 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 1.4 | 7.0 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 1.4 | 4.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 1.3 | 4.0 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 1.3 | 3.9 | GO:0072554 | blood vessel lumenization(GO:0072554) negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.3 | 6.3 | GO:0032901 | positive regulation of neurotrophin production(GO:0032901) |
| 1.2 | 8.3 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 1.2 | 5.8 | GO:0061198 | fungiform papilla development(GO:0061196) fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 1.1 | 3.4 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 1.1 | 9.0 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 1.1 | 6.6 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 1.1 | 5.3 | GO:0046864 | retinol transport(GO:0034633) nose morphogenesis(GO:0043585) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 1.1 | 3.2 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 1.0 | 8.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.0 | 3.0 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.9 | 9.4 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.9 | 5.6 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.9 | 3.7 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.9 | 1.8 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.9 | 4.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.9 | 2.7 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.9 | 5.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.9 | 2.6 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.8 | 13.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.8 | 0.8 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.8 | 5.8 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.8 | 4.0 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.8 | 3.1 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.8 | 8.5 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.7 | 9.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.7 | 2.2 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) positive regulation of pancreatic juice secretion(GO:0090187) positive regulation of somatostatin secretion(GO:0090274) |
| 0.7 | 2.9 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.7 | 7.2 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.7 | 2.1 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.7 | 5.0 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.7 | 4.3 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.7 | 0.7 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.7 | 4.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.7 | 5.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.7 | 6.9 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.7 | 3.4 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.7 | 9.5 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.7 | 2.7 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.7 | 6.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.7 | 2.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.7 | 2.0 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.6 | 2.6 | GO:0097694 | leptotene(GO:0000237) establishment of RNA localization to telomere(GO:0097694) |
| 0.6 | 5.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.6 | 0.6 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.6 | 3.8 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.6 | 1.9 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.6 | 2.5 | GO:0060486 | Clara cell differentiation(GO:0060486) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 | 3.6 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.6 | 2.4 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.6 | 1.8 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.6 | 1.7 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.6 | 2.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.6 | 2.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.5 | 1.1 | GO:0061744 | motor behavior(GO:0061744) |
| 0.5 | 2.2 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.5 | 2.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.5 | 9.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.5 | 3.7 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.5 | 1.0 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.5 | 1.5 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.5 | 4.5 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.5 | 2.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.5 | 2.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.5 | 5.4 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.5 | 2.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.5 | 13.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.5 | 3.4 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.5 | 1.9 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.5 | 8.2 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.5 | 4.8 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.5 | 1.4 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.5 | 1.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.5 | 7.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.5 | 2.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.4 | 2.2 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.4 | 5.8 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.4 | 5.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.4 | 3.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.4 | 1.3 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.4 | 1.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.4 | 1.6 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.4 | 1.2 | GO:0097278 | complement-dependent cytotoxicity(GO:0097278) |
| 0.4 | 4.0 | GO:0032048 | cardiolipin metabolic process(GO:0032048) |
| 0.4 | 6.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.4 | 2.0 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.4 | 1.6 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.4 | 3.0 | GO:2000301 | myeloid progenitor cell differentiation(GO:0002318) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 | 3.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.4 | 3.0 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.4 | 5.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.4 | 1.8 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.4 | 3.2 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.4 | 1.1 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.3 | 1.4 | GO:0061743 | motor learning(GO:0061743) |
| 0.3 | 1.7 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 | 1.4 | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) |
| 0.3 | 1.0 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.3 | 2.4 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.3 | 7.3 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.3 | 7.9 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.3 | 3.3 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 12.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 1.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 1.0 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 1.0 | GO:1990927 | negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.3 | 2.9 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.3 | 1.6 | GO:0019530 | taurine metabolic process(GO:0019530) |
| 0.3 | 7.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.3 | 1.9 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 5.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 0.6 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.3 | 1.2 | GO:2000668 | dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.3 | 1.5 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 | 2.6 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.3 | 3.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.3 | 1.4 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.3 | 6.3 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.3 | 0.6 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.3 | 1.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 5.5 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.3 | 2.4 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.3 | 12.8 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.3 | 3.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.3 | 1.3 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.3 | 1.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.3 | 1.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.3 | 1.0 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.3 | 7.9 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.3 | 0.5 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.3 | 1.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 | 1.5 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 | 4.9 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.2 | 0.7 | GO:1903048 | regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.2 | 1.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.2 | 3.1 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.2 | 3.8 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 1.0 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 | 1.4 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.2 | 1.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 9.7 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 4.7 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.2 | 3.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 2.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.2 | 1.3 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.2 | 0.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.3 | GO:0098543 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.2 | 3.0 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 1.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.2 | 2.5 | GO:0051492 | regulation of actin filament bundle assembly(GO:0032231) regulation of stress fiber assembly(GO:0051492) |
| 0.2 | 1.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.2 | 1.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 1.6 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.2 | 4.5 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.2 | 2.7 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 0.4 | GO:1903431 | positive regulation of neuron maturation(GO:0014042) positive regulation of cell maturation(GO:1903431) |
| 0.2 | 1.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.2 | 6.9 | GO:0007588 | excretion(GO:0007588) |
| 0.2 | 0.8 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.2 | 1.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 1.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 5.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 0.8 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.2 | 4.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 4.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.7 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.2 | 2.2 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.2 | 1.8 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.2 | 1.9 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.2 | 2.1 | GO:1903963 | arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 0.5 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 0.5 | GO:0002842 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.2 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 1.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.2 | 1.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 1.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 3.1 | GO:0032757 | positive regulation of interleukin-8 production(GO:0032757) |
| 0.2 | 2.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.0 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.2 | 0.5 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.8 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.2 | 1.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 11.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 0.6 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.2 | 0.9 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.2 | 0.9 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.2 | 1.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.9 | GO:0048597 | post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.3 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.1 | 1.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 1.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 1.6 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.1 | 0.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 1.7 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.3 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 2.3 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 2.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.9 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 1.1 | GO:1900193 | regulation of oocyte development(GO:0060281) regulation of oocyte maturation(GO:1900193) |
| 0.1 | 1.1 | GO:0021924 | cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.1 | 1.5 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 4.0 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 2.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.4 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 1.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.2 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.1 | 2.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 1.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.0 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 9.3 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.1 | 2.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.8 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 1.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.9 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 1.0 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 0.4 | GO:0070092 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.1 | 0.5 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 | 1.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 1.1 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 1.9 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.1 | 4.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 1.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 1.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.2 | GO:0048070 | regulation of melanocyte differentiation(GO:0045634) regulation of developmental pigmentation(GO:0048070) regulation of pigment cell differentiation(GO:0050932) |
| 0.1 | 2.3 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.1 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 1.4 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) observational learning(GO:0098597) learned vocalization behavior or vocal learning(GO:0098598) |
| 0.1 | 0.5 | GO:0090005 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 1.3 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 1.3 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 1.8 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.1 | 2.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.6 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.1 | 0.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.1 | GO:0045875 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) negative regulation of sister chromatid cohesion(GO:0045875) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.1 | 1.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.0 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 0.4 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 2.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 3.5 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.5 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 2.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.5 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.1 | 0.6 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 0.2 | GO:0070973 | COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 2.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.5 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.1 | 0.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.1 | 2.3 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.1 | 0.3 | GO:0090282 | trophectodermal cell proliferation(GO:0001834) positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.1 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.1 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 2.3 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 0.2 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 1.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 2.1 | GO:0035904 | aorta development(GO:0035904) |
| 0.1 | 1.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 2.4 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 0.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 4.2 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 2.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 3.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 2.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.6 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.1 | 0.4 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.1 | 2.1 | GO:0017145 | stem cell division(GO:0017145) |
| 0.1 | 0.4 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 5.0 | GO:0044782 | cilium organization(GO:0044782) |
| 0.0 | 2.7 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.6 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 1.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 2.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 1.5 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 2.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.7 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.2 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:0090335 | regulation of brown fat cell differentiation(GO:0090335) positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 2.8 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.8 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.2 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.4 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 0.9 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 2.3 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 1.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0061325 | extraocular skeletal muscle development(GO:0002074) cardiac neural crest cell migration involved in outflow tract morphogenesis(GO:0003253) pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) left lung development(GO:0060459) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) endodermal digestive tract morphogenesis(GO:0061031) cardiac neural crest cell differentiation involved in heart development(GO:0061307) cardiac neural crest cell development involved in heart development(GO:0061308) cardiac neural crest cell development involved in outflow tract morphogenesis(GO:0061309) cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.0 | 0.5 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 1.0 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 2.9 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.7 | GO:0044843 | cell cycle G1/S phase transition(GO:0044843) |
| 0.0 | 0.1 | GO:0035405 | negative regulation of B cell proliferation(GO:0030889) histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.8 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 1.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.0 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.8 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.5 | GO:0036125 | mitochondrial fatty acid beta-oxidation multienzyme complex(GO:0016507) fatty acid beta-oxidation multienzyme complex(GO:0036125) |
| 2.6 | 10.3 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 2.5 | 7.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 2.2 | 10.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.9 | 17.4 | GO:0030478 | actin cap(GO:0030478) |
| 1.8 | 7.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 1.7 | 8.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.6 | 6.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 1.2 | 3.5 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.9 | 3.7 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.8 | 8.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.8 | 2.3 | GO:0000801 | central element(GO:0000801) |
| 0.7 | 2.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.7 | 4.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.6 | 3.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.6 | 3.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.6 | 2.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.5 | 7.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.5 | 1.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.4 | 5.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.4 | 2.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.4 | 3.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.4 | 9.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.4 | 3.3 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.4 | 4.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.4 | 2.8 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.4 | 17.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.4 | 12.7 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.4 | 2.6 | GO:0070187 | telosome(GO:0070187) |
| 0.4 | 2.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.4 | 2.5 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.3 | 4.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 3.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.3 | 1.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.3 | 4.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.3 | 1.0 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.3 | 5.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 2.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 3.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 6.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.3 | 6.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 2.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 4.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 3.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 19.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.3 | 0.8 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.3 | 1.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 5.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 7.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 3.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 1.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.2 | 3.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 1.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 0.7 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 6.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 9.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 8.1 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 3.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 1.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 1.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 1.4 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.2 | 0.8 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 5.6 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 2.9 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 7.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.2 | 1.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 10.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 3.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 2.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 5.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 2.1 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 2.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 10.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 5.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 1.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 4.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 7.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 11.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 6.5 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 1.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 3.2 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 7.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.5 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 7.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 2.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 7.2 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 2.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 9.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 5.5 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 2.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 3.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.1 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.1 | 1.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 3.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 3.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 3.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 2.9 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 2.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 1.8 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 2.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 5.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.4 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 6.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 3.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 52.5 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 4.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.5 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.2 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) ribonucleoprotein complex(GO:1990904) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.2 | 21.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 4.1 | 16.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 2.9 | 11.5 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 2.5 | 7.6 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 2.2 | 10.8 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 2.0 | 8.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.9 | 22.5 | GO:0050693 | LBD domain binding(GO:0050693) |
| 1.5 | 5.8 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 1.4 | 16.9 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 1.2 | 6.0 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 1.2 | 3.5 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.1 | 12.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 1.0 | 3.9 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 1.0 | 3.9 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 1.0 | 9.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.9 | 3.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.9 | 8.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.9 | 2.7 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.9 | 3.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.8 | 2.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.8 | 4.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.8 | 6.9 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.8 | 2.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.8 | 5.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.7 | 4.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.7 | 3.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.7 | 2.7 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.7 | 4.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.7 | 5.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.7 | 4.6 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.6 | 3.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.6 | 2.6 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.6 | 15.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.6 | 3.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.6 | 4.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.6 | 5.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.6 | 1.8 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.6 | 1.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.6 | 2.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.5 | 3.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.5 | 6.0 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.5 | 3.7 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.5 | 5.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 2.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.5 | 2.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.5 | 1.9 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.5 | 8.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.5 | 1.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.5 | 4.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.5 | 1.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.4 | 3.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.4 | 2.4 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.4 | 1.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.4 | 14.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.4 | 1.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.4 | 6.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.4 | 9.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.4 | 7.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.4 | 1.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.4 | 8.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 4.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 5.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 3.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 1.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 1.0 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.3 | 1.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.3 | 1.9 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.3 | 6.0 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 4.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 9.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.3 | 2.2 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.3 | 0.8 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.3 | 4.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 1.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.3 | 1.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.3 | 1.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.3 | 0.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.3 | 2.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 4.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.2 | 1.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 1.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 5.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 1.7 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 1.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 4.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 7.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 0.6 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.2 | 4.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 1.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 0.8 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.2 | 1.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 0.6 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.2 | 1.8 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 1.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 3.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.2 | 1.5 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 2.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 3.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 1.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 1.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 3.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 2.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 5.3 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.2 | 1.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.2 | 4.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 2.1 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.2 | 1.0 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 1.5 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.2 | 3.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 7.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 17.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 4.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 2.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 0.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 1.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 2.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 4.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 5.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 4.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.3 | GO:0046977 | TAP binding(GO:0046977) |
| 0.1 | 0.7 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 2.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.8 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.7 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 3.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 2.2 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 2.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 2.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 9.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 19.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 1.0 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 5.6 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 3.2 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 1.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 3.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 2.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 23.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.3 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 4.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 0.3 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.1 | 0.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 3.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.2 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.1 | 1.5 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.1 | 3.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 3.1 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.1 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 25.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.1 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 2.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.5 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 5.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 15.6 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 2.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.9 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 2.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 9.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 6.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 2.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.0 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.4 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 2.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 1.0 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 2.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.5 | GO:0031702 | angiotensin receptor binding(GO:0031701) type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 1.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 2.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 3.9 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 5.2 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 2.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 2.9 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.2 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.8 | 10.2 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.4 | 17.1 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.4 | 10.7 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.3 | 2.1 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.3 | 12.2 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 4.0 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 3.8 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 9.2 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 3.7 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 2.1 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 16.2 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.2 | 5.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.2 | 14.1 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 3.4 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 9.8 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 5.5 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.2 | 5.9 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.2 | 3.8 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.2 | 6.6 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.2 | 6.3 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 1.9 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.2 | 1.9 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.2 | 0.6 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 12.2 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.1 | 2.8 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 6.4 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 6.6 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 3.5 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.1 | 6.8 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.1 | 7.6 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.1 | 3.7 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.1 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.1 | 2.6 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 6.3 | PID_AVB3_INTEGRIN_PATHWAY | Integrins in angiogenesis |
| 0.1 | 2.4 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 11.8 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.4 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 11.5 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 0.9 | PID_ERBB1_DOWNSTREAM_PATHWAY | ErbB1 downstream signaling |
| 0.1 | 1.9 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.3 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 1.1 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.1 | 0.7 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 2.3 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 4.7 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.8 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.1 | 2.4 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 0.8 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.7 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 1.3 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.1 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 3.4 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.9 | 17.4 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.8 | 4.7 | REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.7 | 11.0 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.7 | 11.5 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.6 | 15.1 | REACTOME_APC_CDC20_MEDIATED_DEGRADATION_OF_NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.6 | 22.5 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.6 | 4.0 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.5 | 11.3 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.5 | 3.9 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 16.4 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.4 | 9.2 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.4 | 9.5 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.4 | 4.0 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 3.8 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 3.4 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.3 | 3.7 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.3 | 4.3 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 5.8 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.2 | 3.4 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 4.6 | REACTOME_ARMS_MEDIATED_ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.2 | 3.7 | REACTOME_INTEGRATION_OF_PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 3.7 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 4.6 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 8.7 | REACTOME_PIP3_ACTIVATES_AKT_SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 0.8 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.2 | 2.6 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 17.3 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 2.9 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 1.1 | REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.2 | 3.2 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 2.1 | REACTOME_MUSCLE_CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 5.6 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 13.4 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 2.4 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 8.3 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.8 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.8 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.1 | 4.4 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.1 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.8 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.1 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.2 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.4 | REACTOME_DESTABILIZATION_OF_MRNA_BY_KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 0.9 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.3 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.5 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.7 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 3.5 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.6 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.7 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 0.4 | REACTOME_THROMBIN_SIGNALLING_THROUGH_PROTEINASE_ACTIVATED_RECEPTORS_PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.1 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.9 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.4 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.2 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.3 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.2 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.4 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.2 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 2.9 | REACTOME_TRANSPORT_OF_MATURE_TRANSCRIPT_TO_CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.5 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.5 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 3.8 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_MRNA_STABILITY_BY_PROTEINS_THAT_BIND_AU_RICH_ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.2 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.9 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.1 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME_N_GLYCAN_TRIMMING_IN_THE_ER_AND_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.4 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME_MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_PDGF | Genes involved in Signaling by PDGF |