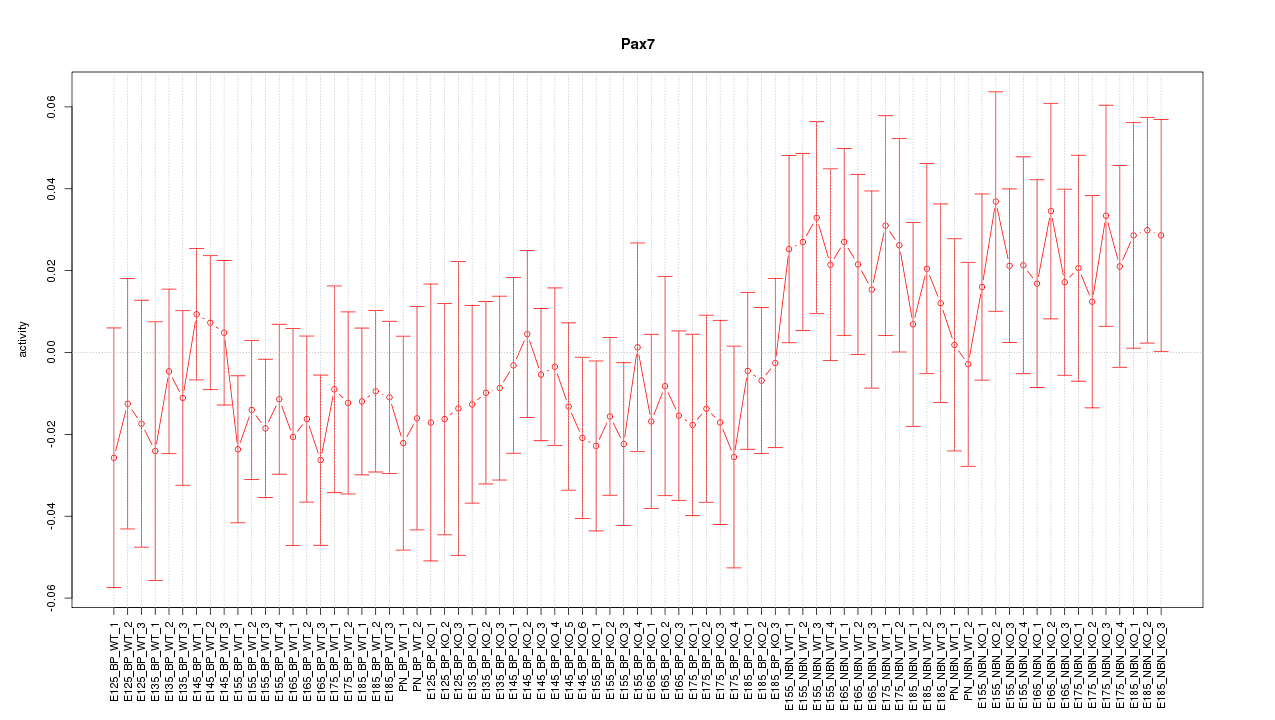
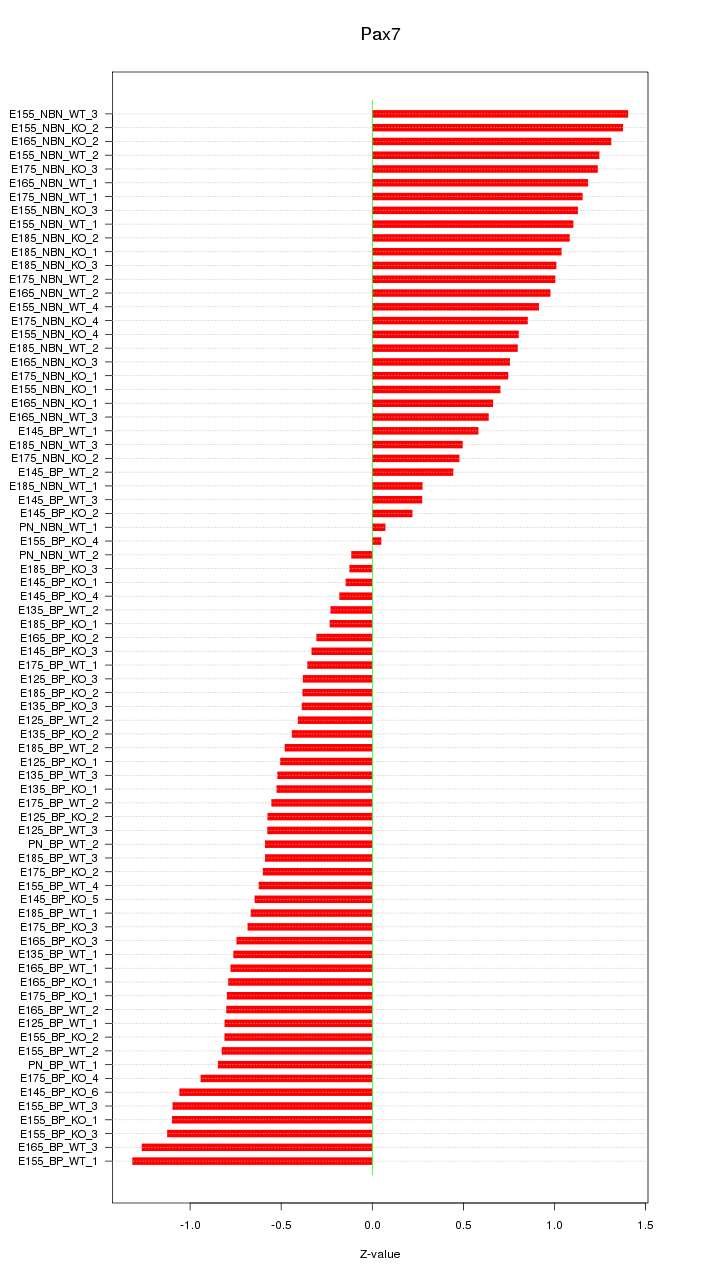
Motif ID: Pax7
Z-value: 0.783
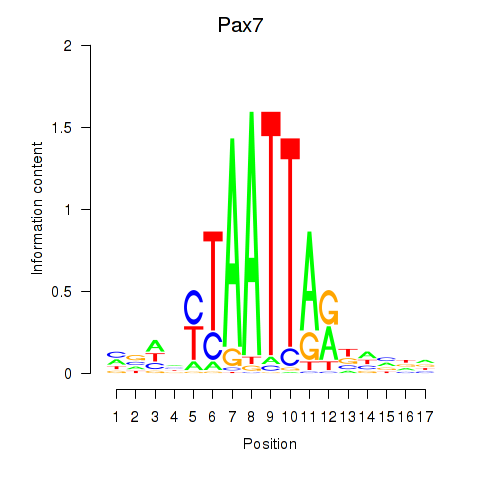
Transcription factors associated with Pax7:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pax7 | ENSMUSG00000028736.7 | Pax7 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax7 | mm10_v2_chr4_-_139833524_139833535 | -0.08 | 4.9e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 35.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.4 | 4.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.9 | 6.6 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.8 | 2.3 | GO:0009838 | abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.7 | 2.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.6 | 7.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 19.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.4 | 4.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.6 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 1.2 | GO:0015791 | polyol transport(GO:0015791) bile acid secretion(GO:0032782) |
| 0.2 | 1.0 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 5.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.2 | 1.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 10.7 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 7.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 5.1 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 | 0.7 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.4 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 7.6 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 1.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 2.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.9 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 1.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 3.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 2.2 | GO:0006364 | rRNA processing(GO:0006364) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.3 | 5.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 7.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 2.3 | GO:0090543 | ESCRT III complex(GO:0000815) Flemming body(GO:0090543) |
| 0.2 | 1.0 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.2 | 2.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 1.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 1.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 10.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 36.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 2.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 19.7 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 5.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 4.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 7.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.4 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 19.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 3.2 | 35.5 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 1.3 | 6.6 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.0 | 5.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.9 | 4.3 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 1.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 4.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 7.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 7.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.4 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 1.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 2.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 5.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 2.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 10.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 4.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 2.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 5.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.7 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 6.6 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 4.2 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.1 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 2.2 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.0 | 2.9 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.8 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.0 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.7 | PID_ATR_PATHWAY | ATR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 19.7 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.5 | 5.7 | REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.4 | 7.6 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 32.8 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 2.2 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.2 | 7.3 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 4.3 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 1.2 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 6.6 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 2.3 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.7 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.1 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |