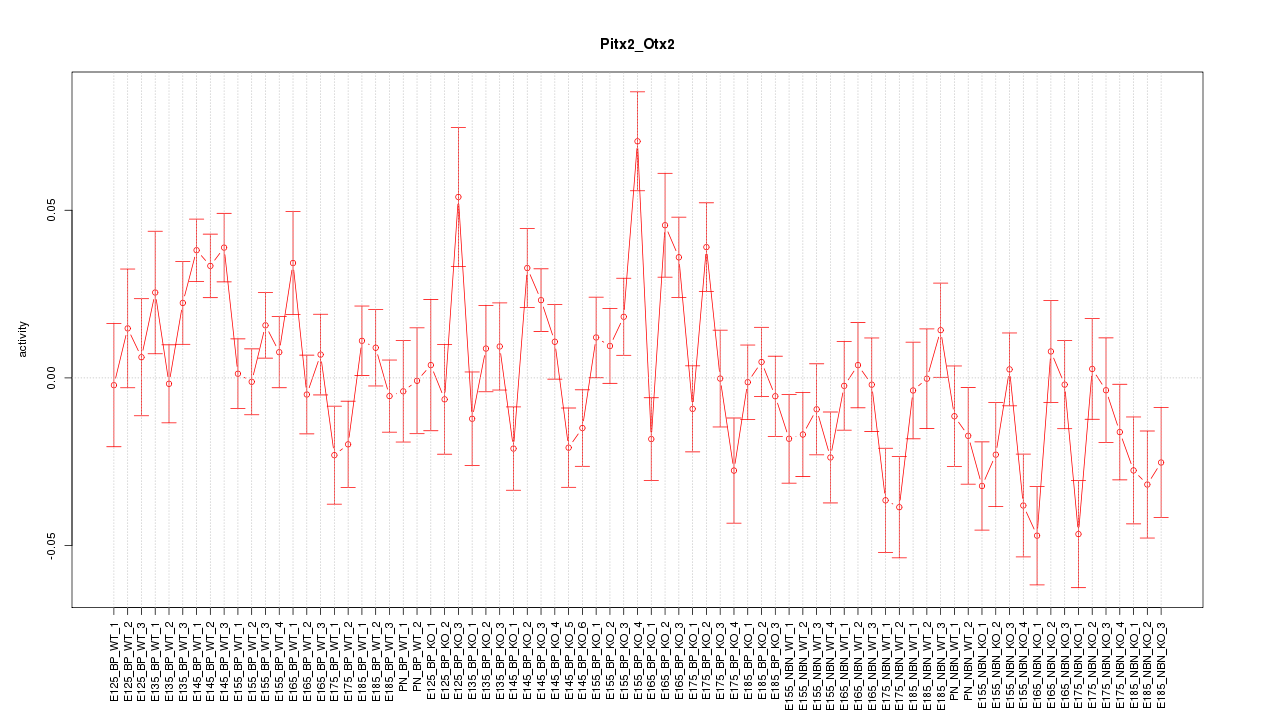
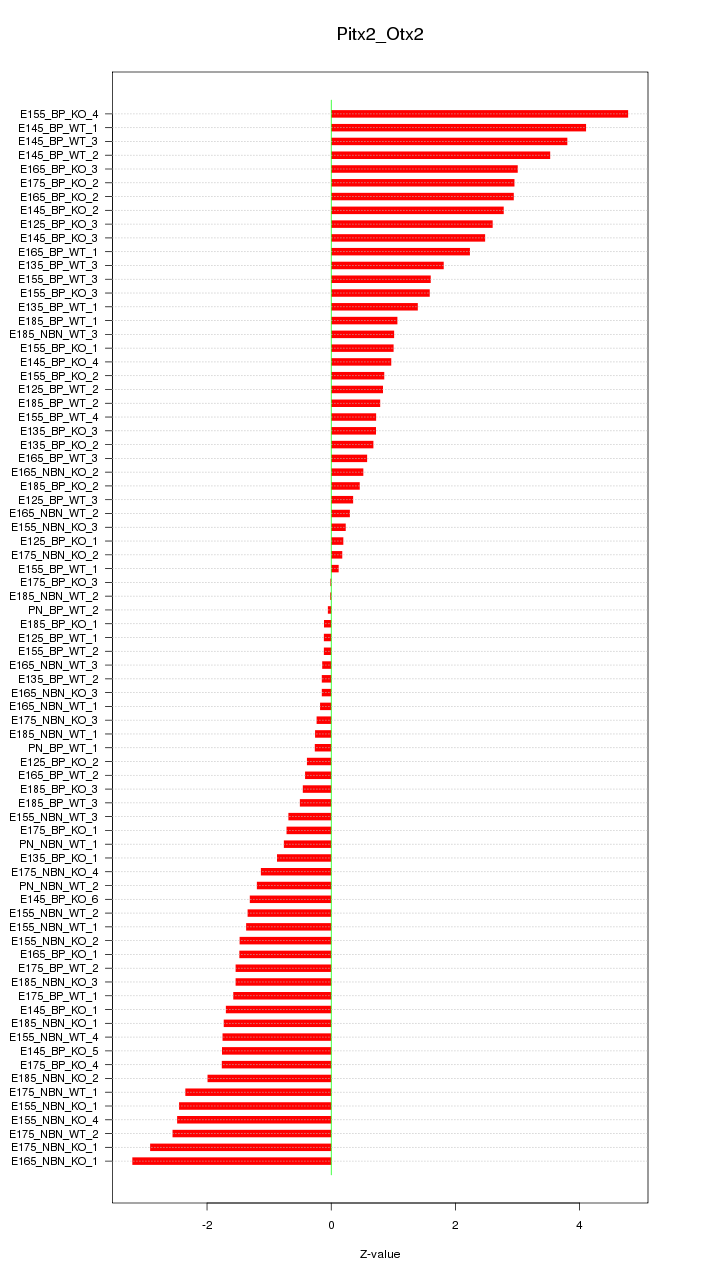
Motif ID: Pitx2_Otx2
Z-value: 1.702
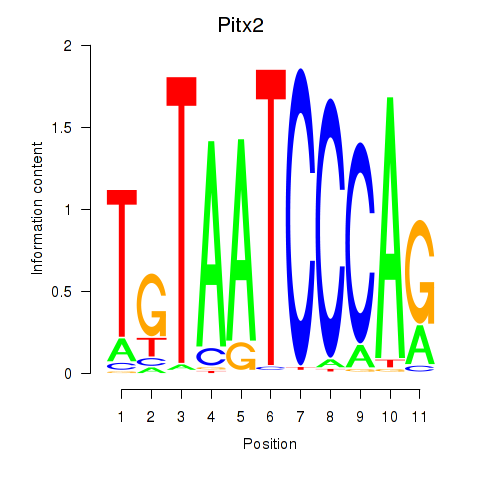
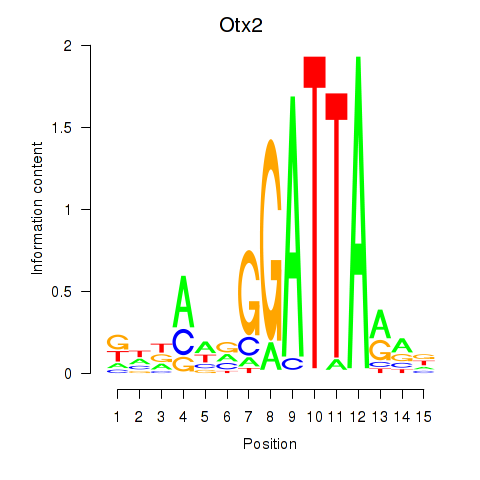
Transcription factors associated with Pitx2_Otx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Otx2 | ENSMUSG00000021848.9 | Otx2 |
| Pitx2 | ENSMUSG00000028023.10 | Pitx2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Otx2 | mm10_v2_chr14_-_48667508_48667644 | 0.19 | 9.5e-02 | Click! |
| Pitx2 | mm10_v2_chr3_+_129199878_129199913 | 0.08 | 5.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) terpenoid catabolic process(GO:0016115) primary alcohol catabolic process(GO:0034310) |
| 3.0 | 8.9 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 2.9 | 8.8 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 2.6 | 13.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 2.3 | 13.9 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 2.3 | 6.8 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 2.0 | 11.9 | GO:0003383 | apical constriction(GO:0003383) |
| 2.0 | 6.0 | GO:0002865 | immune response-inhibiting signal transduction(GO:0002765) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 2.0 | 19.7 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.7 | 5.2 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 1.7 | 5.2 | GO:1902071 | regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 1.6 | 4.9 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 1.6 | 4.8 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 1.6 | 4.8 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 1.6 | 6.2 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 1.6 | 4.7 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 1.6 | 4.7 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.5 | 4.6 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 1.5 | 5.8 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 1.4 | 5.6 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 1.4 | 4.1 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 1.3 | 3.8 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 1.3 | 2.5 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 1.1 | 3.4 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 1.1 | 5.6 | GO:1902995 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 1.1 | 39.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 1.1 | 3.3 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 1.1 | 5.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 1.1 | 3.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 1.1 | 7.6 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 1.1 | 3.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 1.0 | 3.1 | GO:0050787 | glycolate metabolic process(GO:0009441) enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 1.0 | 5.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 1.0 | 2.9 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 1.0 | 6.8 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 1.0 | 2.9 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.0 | 2.9 | GO:1904046 | seryl-tRNA aminoacylation(GO:0006434) negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.9 | 5.7 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.9 | 2.8 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.9 | 4.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.9 | 2.7 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.9 | 2.7 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.9 | 2.7 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.9 | 2.7 | GO:0036233 | glycine import(GO:0036233) |
| 0.9 | 5.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.9 | 3.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.9 | 1.7 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.8 | 2.5 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.8 | 7.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.8 | 2.5 | GO:0006272 | leading strand elongation(GO:0006272) DNA replication proofreading(GO:0045004) |
| 0.8 | 4.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.8 | 5.7 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.8 | 1.6 | GO:1903519 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.8 | 3.9 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.8 | 18.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.8 | 3.9 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.8 | 3.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.8 | 4.5 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.7 | 2.2 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.7 | 3.7 | GO:0061085 | regulation of histone H3-K27 methylation(GO:0061085) positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.7 | 8.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.7 | 2.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.7 | 2.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.7 | 2.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.7 | 2.0 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.7 | 3.4 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.7 | 2.7 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.6 | 2.4 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.6 | 2.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.6 | 2.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.6 | 5.8 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.6 | 2.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.6 | 2.9 | GO:0015817 | histidine transport(GO:0015817) |
| 0.6 | 2.9 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.6 | 1.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.6 | 5.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.6 | 1.7 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.6 | 2.2 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.6 | 4.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.6 | 2.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.5 | 3.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.5 | 6.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.5 | 1.6 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.5 | 3.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.5 | 2.6 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.5 | 1.5 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.5 | 2.5 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.5 | 5.0 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.5 | 1.5 | GO:0036166 | phenotypic switching(GO:0036166) |
| 0.5 | 1.5 | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) |
| 0.5 | 1.9 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.5 | 1.4 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.5 | 1.4 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.5 | 1.4 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.5 | 2.8 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.5 | 1.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.5 | 2.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.4 | 5.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.4 | 0.9 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.4 | 3.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.4 | 2.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 | 1.3 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.4 | 3.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.4 | 2.1 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.4 | 2.5 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.4 | 2.5 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.4 | 6.0 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.4 | 1.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.4 | 3.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.4 | 2.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.4 | 1.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.4 | 2.3 | GO:0036006 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.4 | 1.9 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.4 | 1.9 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.4 | 1.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 3.4 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.4 | 3.0 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.4 | 2.6 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.4 | 3.0 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.4 | 1.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 1.4 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.4 | 0.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.4 | 2.5 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.4 | 2.1 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.4 | 2.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.3 | 3.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.3 | 9.3 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 1.7 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.3 | 3.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.3 | 0.7 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.3 | 1.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.3 | 1.3 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.3 | 1.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.3 | 2.0 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.3 | 1.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 2.5 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 1.0 | GO:0021972 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) regulation of retinal ganglion cell axon guidance(GO:0090259) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.3 | 0.6 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 | 0.9 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.3 | 6.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.3 | 3.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 2.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 2.8 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.3 | 3.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 1.2 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.3 | 3.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.3 | 1.2 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.3 | 3.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.3 | 3.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 | 7.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 7.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.3 | 2.3 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.3 | 0.3 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.3 | 2.5 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.3 | 1.4 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.3 | 1.7 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.3 | 0.8 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.3 | 9.7 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.3 | 1.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.3 | 1.3 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.3 | 1.6 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.3 | 1.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.3 | 1.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 | 2.6 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.3 | 3.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 3.6 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.3 | 2.0 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 0.8 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.3 | 2.0 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.3 | 2.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 0.7 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 1.0 | GO:0080184 | response to stilbenoid(GO:0035634) response to phenylpropanoid(GO:0080184) |
| 0.2 | 1.0 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.2 | 1.0 | GO:0015822 | ornithine transport(GO:0015822) |
| 0.2 | 2.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 1.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.9 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 1.9 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.2 | 1.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.2 | 5.2 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.2 | 1.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 1.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 11.8 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.2 | 1.4 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.2 | 0.9 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 1.6 | GO:0042148 | strand invasion(GO:0042148) |
| 0.2 | 0.9 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.2 | 1.6 | GO:0015675 | nickel cation transport(GO:0015675) |
| 0.2 | 3.4 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.2 | 2.5 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.2 | 2.4 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.2 | 0.9 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.2 | 1.3 | GO:0035878 | nail development(GO:0035878) |
| 0.2 | 2.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 2.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.2 | 3.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 6.3 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.2 | 1.9 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.2 | 1.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 0.8 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.2 | 1.0 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 | 1.4 | GO:0036506 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 2.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 8.9 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.2 | 1.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 0.8 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.2 | 2.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 3.5 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.2 | 1.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 0.6 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.2 | 2.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.5 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.2 | 3.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 0.8 | GO:0002339 | B cell selection(GO:0002339) |
| 0.2 | 2.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 2.8 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.2 | 1.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 0.9 | GO:0015960 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.2 | 6.8 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.2 | 1.3 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.2 | 1.6 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.2 | 2.9 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 0.5 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 | 7.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.2 | 10.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.2 | 0.5 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 | 2.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 0.7 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 4.0 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 3.1 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 2.7 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.2 | 0.5 | GO:0070488 | regulation of integrin biosynthetic process(GO:0045113) neutrophil aggregation(GO:0070488) |
| 0.2 | 1.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.2 | 1.2 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.2 | 1.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.2 | 0.3 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 1.5 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.2 | 1.1 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.2 | 3.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 1.6 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.2 | 17.2 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.2 | 0.8 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 1.8 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.9 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.1 | 1.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.0 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 4.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 1.9 | GO:0071353 | response to interleukin-4(GO:0070670) cellular response to interleukin-4(GO:0071353) |
| 0.1 | 1.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.2 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.1 | 1.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 5.8 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 4.0 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 1.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.3 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 | 0.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 2.9 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.1 | 3.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 | 2.2 | GO:0009261 | purine ribonucleotide catabolic process(GO:0009154) ribonucleotide catabolic process(GO:0009261) |
| 0.1 | 8.6 | GO:1901799 | negative regulation of proteasomal protein catabolic process(GO:1901799) |
| 0.1 | 1.4 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.1 | 4.5 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 1.2 | GO:1901838 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 0.8 | GO:0060325 | head morphogenesis(GO:0060323) face morphogenesis(GO:0060325) |
| 0.1 | 3.3 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.1 | 0.4 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 2.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 2.6 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.1 | 0.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 2.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 2.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.9 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 2.0 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 2.1 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.1 | 0.9 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 1.0 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 4.5 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 1.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 1.2 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 1.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.6 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.1 | 0.8 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 2.1 | GO:0044783 | mitotic G1 DNA damage checkpoint(GO:0031571) G1 DNA damage checkpoint(GO:0044783) |
| 0.1 | 1.4 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 1.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) embryonic brain development(GO:1990403) |
| 0.1 | 2.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 11.7 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 0.2 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 0.9 | GO:0035646 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 0.6 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.8 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.8 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.6 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 2.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 9.7 | GO:0031109 | microtubule polymerization or depolymerization(GO:0031109) |
| 0.1 | 1.0 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.3 | GO:0061303 | cardiac ventricle formation(GO:0003211) soft palate development(GO:0060023) cornea development in camera-type eye(GO:0061303) |
| 0.1 | 1.6 | GO:0071806 | intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.1 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 7.1 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 1.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 1.6 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 0.4 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 0.7 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 2.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 2.5 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 4.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 1.0 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 1.0 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.9 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 4.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 2.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 3.0 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.1 | 0.7 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.1 | 1.0 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.3 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 3.8 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 3.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.8 | GO:0033108 | mitochondrial respiratory chain complex assembly(GO:0033108) |
| 0.1 | 0.5 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 1.8 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 0.9 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.6 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.1 | 0.7 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.6 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.3 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.1 | 0.4 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 1.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 1.0 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.0 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.5 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.6 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 2.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 2.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.3 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 2.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 2.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 1.0 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.1 | 1.5 | GO:1901796 | regulation of signal transduction by p53 class mediator(GO:1901796) |
| 0.1 | 1.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 1.9 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 0.5 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.0 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 2.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.5 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 1.5 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 | 2.5 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.9 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 1.8 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 1.0 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 1.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.5 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.7 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 1.1 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.2 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 1.9 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.1 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 2.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:2000096 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 2.1 | GO:0016331 | morphogenesis of embryonic epithelium(GO:0016331) |
| 0.0 | 0.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.0 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.9 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.6 | GO:0001942 | hair follicle development(GO:0001942) skin epidermis development(GO:0098773) |
| 0.0 | 0.2 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.0 | GO:0048489 | synaptic vesicle transport(GO:0048489) establishment of synaptic vesicle localization(GO:0097480) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 19.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 1.8 | 5.3 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 1.7 | 5.2 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 1.6 | 6.3 | GO:0032021 | NELF complex(GO:0032021) |
| 1.5 | 9.1 | GO:1990393 | 3M complex(GO:1990393) |
| 1.5 | 11.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.4 | 4.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.4 | 9.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.2 | 3.5 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 1.2 | 6.9 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 1.1 | 3.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.1 | 4.3 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 1.1 | 4.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 1.0 | 2.9 | GO:0071914 | prominosome(GO:0071914) |
| 1.0 | 2.9 | GO:1990047 | spindle matrix(GO:1990047) |
| 1.0 | 3.9 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 1.0 | 2.9 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.9 | 3.8 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.9 | 2.8 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.9 | 3.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.8 | 5.0 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.8 | 7.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.8 | 15.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.8 | 4.8 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.8 | 3.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.8 | 6.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.7 | 7.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.7 | 9.9 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.7 | 8.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.6 | 1.9 | GO:0000801 | central element(GO:0000801) |
| 0.6 | 2.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.6 | 43.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.6 | 7.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.6 | 8.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.6 | 2.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.6 | 3.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.6 | 28.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.6 | 5.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.5 | 19.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.5 | 3.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.5 | 1.5 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.5 | 3.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.5 | 1.5 | GO:0000811 | GINS complex(GO:0000811) |
| 0.5 | 2.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 5.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.5 | 4.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.5 | 1.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.5 | 1.9 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.5 | 6.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.5 | 6.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.4 | 0.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.4 | 5.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.4 | 1.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.4 | 1.6 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.4 | 3.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.4 | 12.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.4 | 4.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.4 | 2.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.3 | 4.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 1.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.3 | 3.9 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 1.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 1.3 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.3 | 2.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 2.8 | GO:0045252 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.3 | 2.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.3 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 2.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.3 | 2.7 | GO:0000243 | commitment complex(GO:0000243) |
| 0.3 | 12.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.3 | 1.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 0.5 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.3 | 2.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.3 | 2.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 1.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 1.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.2 | 3.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.4 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 0.9 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.2 | 0.9 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.2 | 5.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 2.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 1.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 1.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 17.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 3.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 1.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 2.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 1.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 3.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 3.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 6.5 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.2 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 1.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 8.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.2 | 2.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 1.7 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.2 | 1.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.2 | 5.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.2 | 1.5 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.2 | 1.8 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.5 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.2 | 0.3 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.2 | 4.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 0.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 3.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.2 | 0.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 1.6 | GO:0046930 | pore complex(GO:0046930) Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 8.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 3.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 2.2 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 1.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 0.5 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 5.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 6.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 1.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 4.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 6.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 6.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 6.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.7 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 7.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 5.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 19.4 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 3.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 29.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 2.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.5 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.1 | 1.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 1.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 3.0 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 1.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.0 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 1.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.9 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 0.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 3.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.6 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 1.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 4.8 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 4.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.6 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 0.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 2.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 5.1 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 2.8 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 7.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 3.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 3.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 2.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.0 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 1.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 1.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.4 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 0.4 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 2.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 18.6 | GO:0005739 | mitochondrion(GO:0005739) |
| 0.0 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0035859 | Seh1-associated complex(GO:0035859) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 2.7 | 40.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 2.1 | 10.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 1.9 | 5.7 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 1.9 | 5.6 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 1.6 | 4.8 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 1.6 | 12.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.6 | 6.2 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 1.5 | 7.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 1.5 | 2.9 | GO:0070905 | serine binding(GO:0070905) |
| 1.4 | 19.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 1.4 | 4.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.3 | 9.1 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.3 | 5.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.3 | 6.3 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 1.2 | 3.7 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 1.2 | 4.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 1.2 | 3.6 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.1 | 3.4 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 1.0 | 3.1 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 1.0 | 5.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 1.0 | 3.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 1.0 | 6.0 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 1.0 | 2.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 1.0 | 2.9 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 1.0 | 4.8 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 1.0 | 5.8 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.0 | 1.0 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.9 | 2.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.9 | 2.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.9 | 2.7 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.9 | 5.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.8 | 2.5 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.8 | 4.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.8 | 5.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.8 | 2.5 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.8 | 3.2 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.8 | 3.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.8 | 4.5 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.8 | 12.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.8 | 2.3 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.8 | 3.0 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.7 | 2.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.7 | 7.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.7 | 5.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.7 | 0.7 | GO:0005402 | cation:sugar symporter activity(GO:0005402) |
| 0.6 | 2.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.6 | 2.4 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.6 | 2.4 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.6 | 1.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.6 | 8.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.6 | 7.7 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.6 | 1.8 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.6 | 2.9 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.6 | 1.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.6 | 2.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.6 | 6.8 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.6 | 1.7 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.6 | 2.8 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.6 | 2.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.5 | 4.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.5 | 3.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.5 | 2.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.5 | 1.5 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.5 | 4.0 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.5 | 14.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.5 | 5.0 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.5 | 1.4 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.5 | 5.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.5 | 2.3 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.5 | 1.4 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.5 | 2.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.4 | 1.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.4 | 2.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 3.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.4 | 3.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 2.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 4.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 6.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 1.2 | GO:0070140 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.4 | 8.9 | GO:0016675 | oxidoreductase activity, acting on a heme group of donors(GO:0016675) |
| 0.4 | 7.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.4 | 1.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 8.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.4 | 1.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.4 | 1.9 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.4 | 66.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.4 | 1.5 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.4 | 2.6 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.4 | 12.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.4 | 1.8 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 1.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.3 | 1.4 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.3 | 1.7 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.3 | 1.4 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.3 | 2.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.3 | 1.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 3.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.0 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.3 | 2.9 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.3 | 0.6 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.3 | 0.9 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.3 | 1.7 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.3 | 2.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 0.8 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.3 | 1.4 | GO:0016531 | metallochaperone activity(GO:0016530) copper chaperone activity(GO:0016531) |
| 0.3 | 3.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 2.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 1.9 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.3 | 2.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 6.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.3 | 0.8 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.3 | 2.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 12.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 5.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 4.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 1.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.2 | 4.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 0.9 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 0.7 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.2 | 3.7 | GO:0016594 | glycine binding(GO:0016594) |
| 0.2 | 1.4 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.2 | 1.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 2.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 1.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 7.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 1.6 | GO:0015099 | nickel cation transmembrane transporter activity(GO:0015099) |
| 0.2 | 4.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 2.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 3.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.2 | 1.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 9.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.2 | 0.9 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 2.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 4.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 1.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 9.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 1.0 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.2 | 1.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 2.8 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 2.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 3.1 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 1.9 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 1.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 2.2 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 9.8 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.2 | 1.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 2.0 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 3.0 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 1.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 2.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.2 | 2.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.2 | 3.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 3.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 2.7 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 0.3 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 1.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 3.8 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 3.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 3.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 2.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.6 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 1.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 5.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.4 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 2.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.2 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 1.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 5.4 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 1.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 13.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 2.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 3.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 1.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 1.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.8 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 3.4 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.6 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.5 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 1.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.3 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.1 | 1.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 8.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 2.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.5 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 1.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.9 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 1.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.6 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 1.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.9 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 2.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.2 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 0.8 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.1 | 0.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 23.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 10.4 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.1 | 2.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.1 | 1.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 2.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 2.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 3.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.4 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.1 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 3.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 1.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 9.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 1.0 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 2.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 2.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 3.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.5 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 1.3 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 1.3 | GO:0016835 | carbon-oxygen lyase activity(GO:0016835) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.7 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 1.6 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 2.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 3.0 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 1.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 1.1 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 2.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 2.0 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.5 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.4 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.8 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.8 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 9.5 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.6 | 18.3 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.6 | 4.1 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.5 | 25.2 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.5 | 2.4 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.4 | 16.5 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.4 | 12.2 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.4 | 3.5 | PID_NFKAPPAB_ATYPICAL_PATHWAY | Atypical NF-kappaB pathway |
| 0.3 | 13.4 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.3 | 5.1 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 2.9 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 4.5 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.2 | 11.7 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 4.8 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.2 | 5.3 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 2.0 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.2 | 5.8 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.2 | 1.9 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.2 | 5.6 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 15.5 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 4.4 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.9 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.5 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.1 | 5.0 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 3.2 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.8 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.1 | PID_AR_NONGENOMIC_PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 1.6 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 3.8 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.8 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.9 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.7 | PID_BARD1_PATHWAY | BARD1 signaling events |
| 0.1 | 2.3 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.1 | 4.4 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 5.2 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.6 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.8 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.7 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.1 | 5.4 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.6 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 2.1 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.4 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.7 | PID_IL23_PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.8 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 0.7 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.1 | 1.3 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.0 | 0.1 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.5 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.0 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 1.9 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.8 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.2 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 5.9 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 5.1 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.3 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 0.7 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.6 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.2 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.2 | PID_NFAT_3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 21.2 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 1.2 | 19.8 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.9 | 19.7 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.8 | 2.5 | REACTOME_SIGNALING_BY_CONSTITUTIVELY_ACTIVE_EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.8 | 4.7 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.7 | 5.2 | REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILLING_BY_DNA_POL_IN_TC_NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.7 | 11.1 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.6 | 70.0 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.6 | 15.7 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.5 | 11.2 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.5 | 4.9 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.5 | 1.5 | REACTOME_HORMONE_LIGAND_BINDING_RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.5 | 27.1 | REACTOME_SCFSKP2_MEDIATED_DEGRADATION_OF_P27_P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.4 | 2.8 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.4 | 2.9 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 4.8 | REACTOME_CYTOKINE_SIGNALING_IN_IMMUNE_SYSTEM | Genes involved in Cytokine Signaling in Immune system |
| 0.3 | 5.5 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 9.6 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.3 | 6.9 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.3 | 18.7 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 16.5 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.3 | 3.6 | REACTOME_GLUCAGON_TYPE_LIGAND_RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.2 | 4.0 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 4.1 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 2.2 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 3.1 | REACTOME_NFKB_ACTIVATION_THROUGH_FADD_RIP1_PATHWAY_MEDIATED_BY_CASPASE_8_AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 2.8 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 6.3 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 11.0 | REACTOME_PHASE_II_CONJUGATION | Genes involved in Phase II conjugation |
| 0.2 | 2.8 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.2 | 17.3 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.2 | 5.6 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 2.3 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 5.2 | REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 3.1 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 2.9 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 6.5 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 4.2 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 5.7 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.2 | 5.7 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 5.5 | REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.2 | 2.2 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.9 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.6 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 12.4 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.6 | REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.1 | 2.6 | REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.8 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.3 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.3 | REACTOME_ELONGATION_ARREST_AND_RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.2 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 5.8 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.5 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 2.6 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 5.6 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.3 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.4 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.1 | 3.8 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 1.4 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.6 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.6 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.3 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.2 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.9 | REACTOME_EXTRACELLULAR_MATRIX_ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 2.2 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.0 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.1 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 3.1 | REACTOME_PRE_NOTCH_EXPRESSION_AND_PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 1.2 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.2 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.6 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.5 | REACTOME_SPHINGOLIPID_METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 5.0 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.7 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 0.9 | REACTOME_PHOSPHORYLATION_OF_THE_APC_C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 0.9 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.5 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.0 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.1 | 1.2 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.4 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.7 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.0 | 2.0 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.5 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.0 | REACTOME_SIGNALING_BY_FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.8 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.8 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME_DESTABILIZATION_OF_MRNA_BY_KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.8 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.8 | REACTOME_TRANSLATION | Genes involved in Translation |
| 0.0 | 0.9 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME_DAG_AND_IP3_SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.5 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.1 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.5 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.4 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.1 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.8 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |