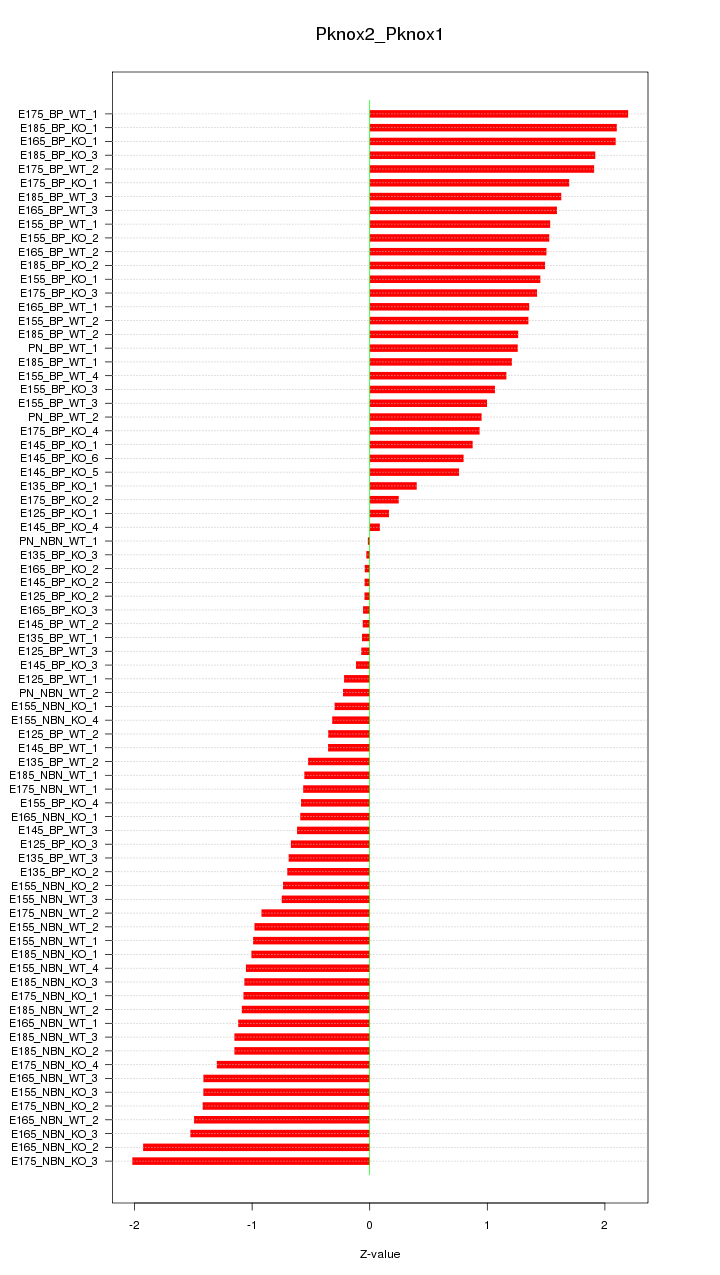
Motif ID: Pknox2_Pknox1
Z-value: 1.114
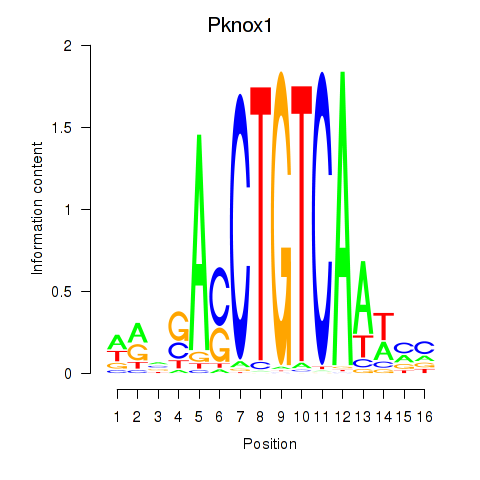
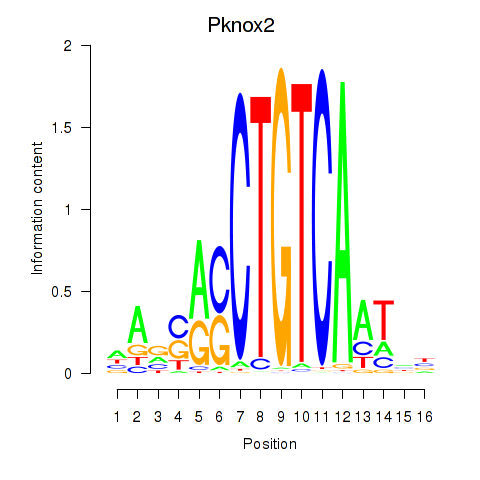
Transcription factors associated with Pknox2_Pknox1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pknox1 | ENSMUSG00000006705.6 | Pknox1 |
| Pknox2 | ENSMUSG00000035934.9 | Pknox2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pknox2 | mm10_v2_chr9_-_37147257_37147323 | 0.30 | 9.1e-03 | Click! |
| Pknox1 | mm10_v2_chr17_+_31564749_31564854 | -0.22 | 4.9e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 3.2 | 9.7 | GO:1901228 | regulation of osteoclast proliferation(GO:0090289) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) negative regulation of bone development(GO:1903011) |
| 2.4 | 11.8 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 1.3 | 7.6 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 1.2 | 8.1 | GO:1901300 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 1.1 | 9.8 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.9 | 6.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.8 | 12.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.7 | 5.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.7 | 2.1 | GO:0070428 | granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.6 | 5.4 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.6 | 2.9 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.6 | 1.7 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.6 | 8.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.5 | 1.6 | GO:0030070 | insulin processing(GO:0030070) |
| 0.5 | 1.5 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.5 | 1.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 4.7 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.5 | 1.4 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.4 | 2.6 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.4 | 0.8 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.4 | 3.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.4 | 2.0 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.4 | 7.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.4 | 4.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.4 | 4.7 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.3 | 1.7 | GO:2000325 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.3 | 2.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 2.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 3.9 | GO:0036315 | cellular response to sterol(GO:0036315) regulation of cholesterol homeostasis(GO:2000188) |
| 0.3 | 2.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.3 | 0.8 | GO:0007621 | negative regulation of female receptivity(GO:0007621) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of cellular pH reduction(GO:0032847) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 0.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 | 1.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 0.7 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.2 | 2.1 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.2 | 1.2 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 1.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 1.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.2 | 0.7 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 3.0 | GO:0006972 | hyperosmotic response(GO:0006972) negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.2 | 2.8 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 0.4 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 1.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.5 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.3 | GO:1904580 | quinolinate biosynthetic process(GO:0019805) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) regulation of intracellular mRNA localization(GO:1904580) |
| 0.1 | 3.5 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 6.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 5.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 2.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 5.6 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 13.1 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.1 | 1.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 1.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 3.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 5.9 | GO:0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) |
| 0.1 | 1.9 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 0.8 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 1.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 5.8 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 1.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.6 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.7 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 2.1 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 3.6 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.7 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 2.3 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 4.1 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0000801 | central element(GO:0000801) |
| 0.7 | 8.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.7 | 2.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 1.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 3.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 2.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 2.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 1.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 5.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 3.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 1.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 7.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.7 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 1.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 1.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 2.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 20.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 3.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 3.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 6.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 7.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 2.0 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 11.4 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 29.3 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 1.5 | 6.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 1.5 | 8.9 | GO:0070728 | leucine binding(GO:0070728) |
| 1.1 | 24.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 1.1 | 9.7 | GO:0036122 | BMP binding(GO:0036122) |
| 1.0 | 3.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.7 | 9.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.6 | 4.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.5 | 1.5 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.5 | 4.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.4 | 8.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.4 | 1.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 3.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 2.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 3.0 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.3 | 0.8 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.2 | 0.7 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.2 | 0.8 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.2 | 1.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 4.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 1.6 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.2 | 1.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 2.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 2.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 6.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.4 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 8.2 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 2.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 27.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 1.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 5.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 4.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 3.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 2.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 6.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 6.3 | GO:0003712 | transcription cofactor activity(GO:0003712) |
| 0.0 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.2 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 8.1 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 9.7 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.2 | 4.5 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 12.6 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.6 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 5.8 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 11.5 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 8.9 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.1 | 2.1 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 9.2 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.1 | 3.0 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.1 | 3.9 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 1.7 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.0 | 4.4 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.3 | PID_PLK1_PATHWAY | PLK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.8 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.6 | 9.8 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 12.6 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.4 | 8.1 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 4.0 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 3.2 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 6.1 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 3.9 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 4.5 | REACTOME_DOWNSTREAM_TCR_SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 0.8 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.0 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.1 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.9 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.4 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.7 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.6 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.8 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.3 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 5.8 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.7 | REACTOME_GLUCOSE_METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 3.2 | REACTOME_GLYCEROPHOSPHOLIPID_BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.7 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.8 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.0 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |