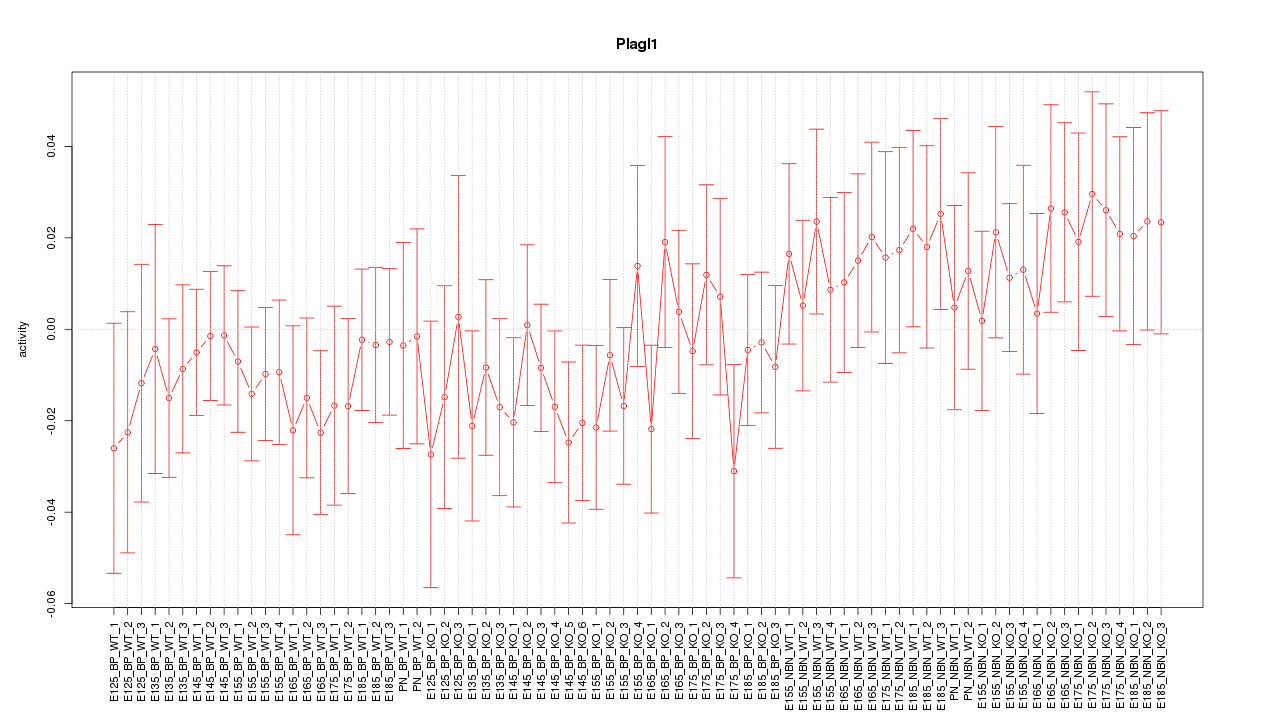
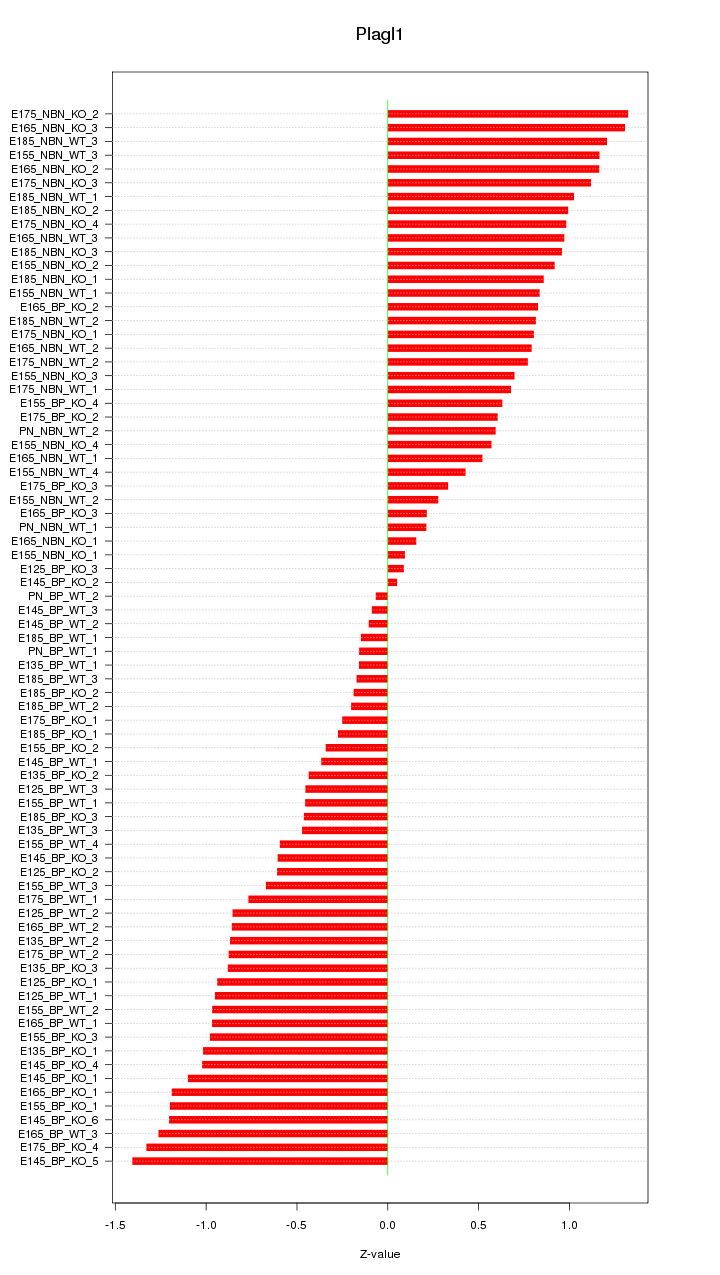
Motif ID: Plagl1
Z-value: 0.785
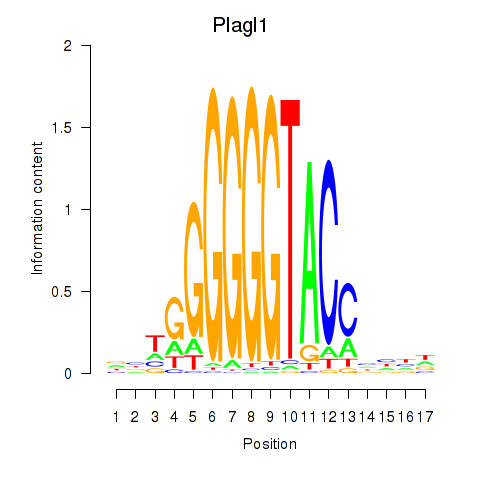
Transcription factors associated with Plagl1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Plagl1 | ENSMUSG00000019817.12 | Plagl1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Plagl1 | mm10_v2_chr10_+_13090788_13090843 | -0.60 | 6.1e-09 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 2.3 | 22.6 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 2.0 | 8.0 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 1.1 | 3.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 1.1 | 7.8 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.9 | 4.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.7 | 2.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.5 | 9.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.5 | 6.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.5 | 5.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 1.8 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.4 | 4.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.4 | 4.8 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.4 | 14.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 | 6.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.3 | 5.4 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.3 | 0.8 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.2 | 2.4 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.2 | 2.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 1.4 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.2 | 2.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 8.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 2.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 1.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 2.3 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.2 | 1.8 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.2 | 1.0 | GO:0060744 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.2 | 1.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 2.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.7 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 4.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 3.0 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.3 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.5 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.6 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.4 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 1.9 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 3.5 | GO:0070542 | response to fatty acid(GO:0070542) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.5 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 1.4 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 | 2.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 3.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 2.1 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.1 | 3.7 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 3.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.3 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.9 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.0 | 0.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) negative regulation of trophoblast cell migration(GO:1901164) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 3.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.0 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 4.8 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 1.3 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.2 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 1.6 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 1.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.0 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 3.4 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.4 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 2.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 11.5 | GO:0007155 | cell adhesion(GO:0007155) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.5 | 6.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 4.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 3.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.4 | 1.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.3 | 7.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 6.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 17.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 2.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 1.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 22.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 8.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 3.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 1.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 3.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 7.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 4.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 4.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 5.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 4.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 3.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 2.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 2.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 3.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 3.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 2.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 4.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.6 | GO:0031966 | mitochondrial membrane(GO:0031966) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 2.7 | 8.0 | GO:0016015 | morphogen activity(GO:0016015) |
| 1.9 | 7.8 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 1.5 | 4.6 | GO:0070905 | serine binding(GO:0070905) |
| 0.9 | 3.5 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.8 | 9.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.8 | 5.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.7 | 2.2 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.7 | 2.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.6 | 6.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.4 | 2.6 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.4 | 2.3 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 4.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 1.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.3 | 0.9 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 2.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.3 | 1.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.3 | 22.6 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.2 | 0.9 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 6.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 3.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 1.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 5.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.5 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 2.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 4.9 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 1.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 19.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 17.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 2.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 4.5 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 1.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.4 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 3.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 21.2 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 7.4 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 8.0 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 2.3 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.1 | 6.1 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.4 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.1 | 6.9 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.1 | 1.8 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 8.8 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.3 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.3 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.1 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 2.3 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 22.6 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.7 | 5.4 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 4.8 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 4.6 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 7.9 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 6.1 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 14.5 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.9 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 3.6 | REACTOME_PLC_BETA_MEDIATED_EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 1.8 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 8.0 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.4 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PLC_BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 1.8 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.9 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 2.4 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.4 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 2.2 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.5 | REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_LATE_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME_EXTRACELLULAR_MATRIX_ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.7 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.0 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |