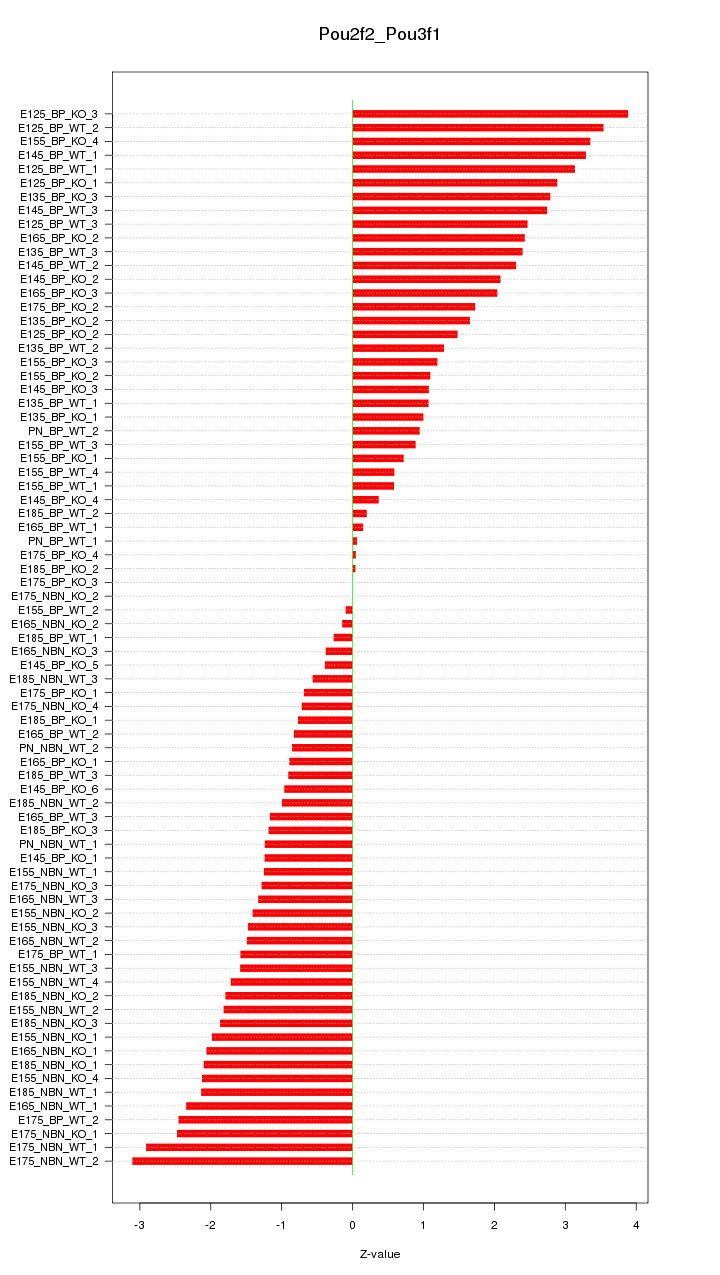
Motif ID: Pou2f2_Pou3f1
Z-value: 1.741

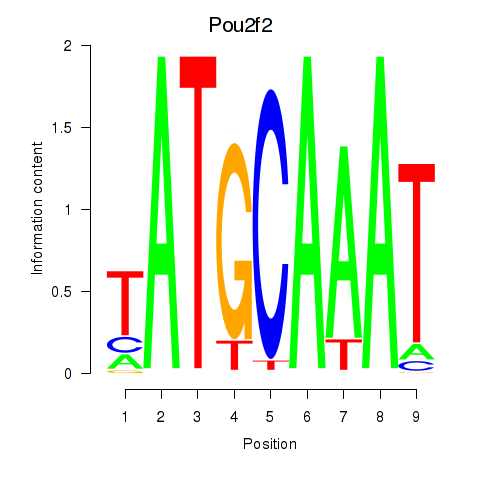
Transcription factors associated with Pou2f2_Pou3f1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pou2f2 | ENSMUSG00000008496.12 | Pou2f2 |
| Pou3f1 | ENSMUSG00000090125.2 | Pou3f1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f1 | mm10_v2_chr4_+_124657646_124657656 | -0.55 | 1.7e-07 | Click! |
| Pou2f2 | mm10_v2_chr7_-_25132473_25132512 | -0.26 | 2.2e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 26.6 | GO:0060853 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 7.1 | 28.4 | GO:0021648 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) cranial nerve formation(GO:0021603) vestibulocochlear nerve morphogenesis(GO:0021648) |
| 6.1 | 18.3 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 5.8 | 29.1 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 5.1 | 15.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 4.9 | 54.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 4.7 | 61.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 4.5 | 9.1 | GO:0061344 | regulation of cell adhesion involved in heart morphogenesis(GO:0061344) positive regulation of ephrin receptor signaling pathway(GO:1901189) |
| 3.9 | 47.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 3.7 | 7.4 | GO:0021546 | rhombomere development(GO:0021546) |
| 3.0 | 11.9 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 2.9 | 11.8 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 2.9 | 49.5 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 2.8 | 8.5 | GO:1904170 | regulation of bleb assembly(GO:1904170) |
| 2.5 | 9.9 | GO:0070269 | pyroptosis(GO:0070269) |
| 2.3 | 13.9 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 2.2 | 25.8 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 2.1 | 8.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 2.0 | 5.9 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 1.9 | 5.8 | GO:0030539 | male genitalia development(GO:0030539) |
| 1.7 | 29.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 1.7 | 5.2 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 1.7 | 3.3 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 1.6 | 11.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 1.5 | 19.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 1.5 | 4.4 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 1.5 | 4.4 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 1.4 | 12.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 1.3 | 26.0 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 1.3 | 19.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.3 | 8.8 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 1.2 | 34.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 1.2 | 8.4 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 1.2 | 3.5 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 1.2 | 9.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 1.2 | 8.1 | GO:0003263 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) |
| 1.1 | 9.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 1.0 | 7.9 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.9 | 2.7 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.9 | 2.6 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.9 | 7.9 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.8 | 10.9 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.7 | 10.4 | GO:0072189 | ureter development(GO:0072189) |
| 0.7 | 2.1 | GO:0070602 | regulation of chondrocyte development(GO:0061181) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.7 | 2.8 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.6 | 5.1 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.6 | 3.0 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.6 | 1.8 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.6 | 63.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.6 | 3.9 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.5 | 3.7 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.5 | 23.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.5 | 2.0 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.5 | 24.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.5 | 1.9 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.5 | 4.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.4 | 0.9 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.4 | 3.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.4 | 6.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 2.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.4 | 2.8 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.4 | 2.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 3.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.3 | 9.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.3 | 15.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 3.7 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.3 | 1.0 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.3 | 2.0 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 3.4 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.3 | 2.2 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.3 | 2.8 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.3 | 29.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.3 | 1.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.3 | 12.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.2 | 8.4 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.2 | 1.6 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 0.9 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.2 | 6.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 3.8 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 12.4 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.2 | 4.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.2 | 1.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.2 | 1.9 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.2 | 0.7 | GO:0060800 | astrocyte fate commitment(GO:0060018) regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 8.1 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.2 | 0.5 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.1 | 4.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 2.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 6.2 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.1 | 0.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.5 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 1.8 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 2.5 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.1 | 3.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 2.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 2.8 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.1 | 7.5 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 2.9 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 0.7 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 2.9 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 6.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.1 | 4.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 3.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 3.7 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.0 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 1.6 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 6.6 | GO:0021700 | developmental maturation(GO:0021700) |
| 0.0 | 4.2 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.3 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 29.5 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 4.2 | 20.9 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 3.4 | 23.8 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 2.5 | 9.9 | GO:0061702 | inflammasome complex(GO:0061702) |
| 2.2 | 53.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.7 | 8.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.5 | 4.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 1.1 | 4.2 | GO:0008623 | CHRAC(GO:0008623) |
| 1.0 | 73.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.8 | 10.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.8 | 9.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.8 | 3.1 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.7 | 12.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.6 | 3.8 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.5 | 2.7 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.4 | 46.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.4 | 9.9 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.4 | 5.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.4 | 2.4 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.4 | 4.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.4 | 26.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 6.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.3 | 14.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.3 | 2.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 17.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 19.2 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 0.6 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 63.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.2 | 5.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 9.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 52.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.2 | 9.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 4.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 10.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 6.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 15.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 6.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 8.8 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 2.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 11.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 146.8 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.7 | 32.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 3.3 | 52.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 2.4 | 11.9 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 2.3 | 13.9 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 2.2 | 10.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 1.4 | 9.9 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 1.3 | 7.9 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 1.3 | 11.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.3 | 8.9 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 1.1 | 4.4 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 1.0 | 23.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 1.0 | 6.8 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.9 | 12.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.9 | 14.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.8 | 179.6 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.8 | 37.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.7 | 3.0 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.7 | 7.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.5 | 3.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.5 | 2.0 | GO:0043426 | MRF binding(GO:0043426) |
| 0.5 | 2.9 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.5 | 8.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 111.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.4 | 4.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 24.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.4 | 8.6 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.4 | 12.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.3 | 3.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 5.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.3 | 2.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.3 | 4.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.3 | 3.7 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 3.9 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.3 | 2.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 2.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.2 | 5.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 1.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.2 | 2.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 4.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 6.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.2 | 2.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 2.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 2.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 4.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 2.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 2.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 2.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 13.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 3.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 4.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 33.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.1 | 2.7 | GO:0005507 | copper ion binding(GO:0005507) beta-tubulin binding(GO:0048487) |
| 0.1 | 4.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 4.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 7.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.2 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 7.0 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 1.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin binding(GO:0030332) |
| 0.0 | 5.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.9 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 3.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 12.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 36.2 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 4.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 61.0 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 1.9 | 69.0 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.8 | 44.0 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.5 | 21.7 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.4 | 25.5 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.4 | 24.7 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.4 | 25.7 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.4 | 19.3 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.4 | 11.9 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.3 | 28.6 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.3 | 11.3 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.3 | 4.2 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.2 | 9.8 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.2 | 12.2 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.2 | 4.0 | PID_IL23_PATHWAY | IL23-mediated signaling events |
| 0.1 | 4.3 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.5 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.7 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.8 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.1 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 3.7 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 20.3 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 3.0 | 23.8 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 2.3 | 84.3 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 2.1 | 29.1 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 1.5 | 24.7 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 1.1 | 14.7 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.8 | 11.8 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.8 | 19.3 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.6 | 8.4 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.6 | 10.2 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 2.6 | REACTOME_ELEVATION_OF_CYTOSOLIC_CA2_LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.4 | 16.7 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 10.9 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 8.8 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.2 | 15.1 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 4.9 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 2.8 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.2 | 22.9 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 13.5 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 4.4 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.0 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.6 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 15.6 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.2 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.1 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.3 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |