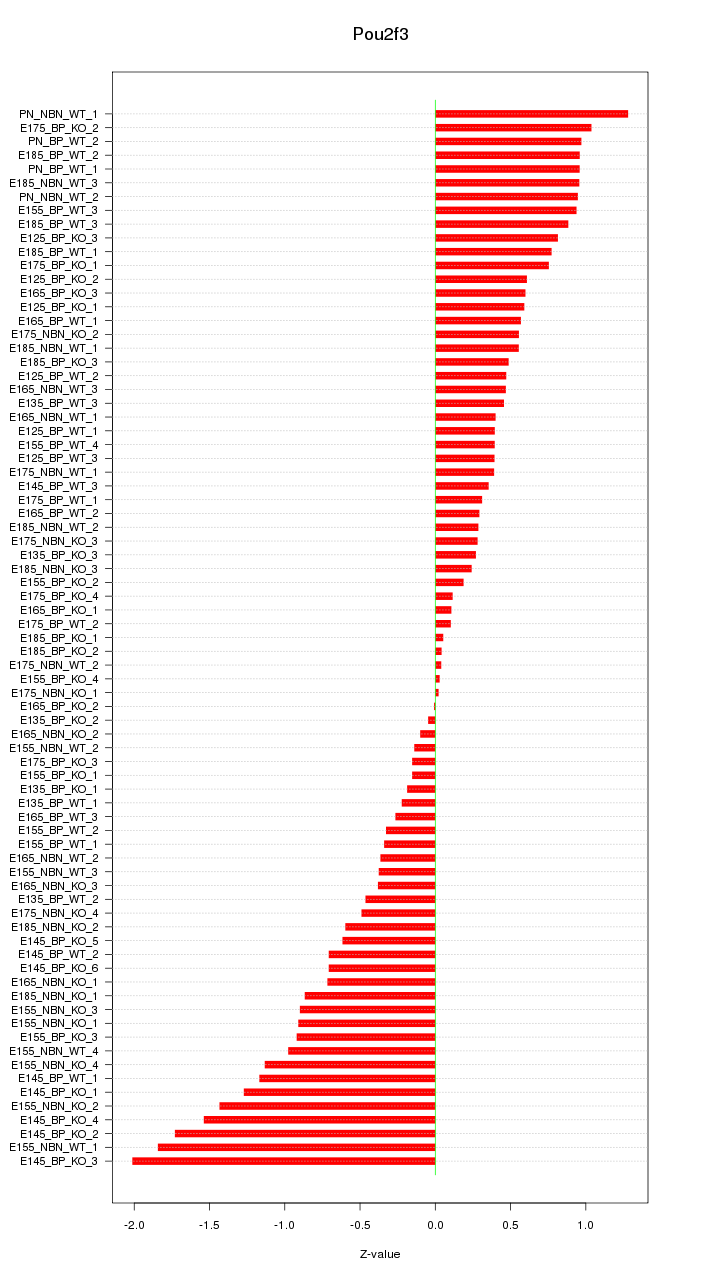
Motif ID: Pou2f3
Z-value: 0.739
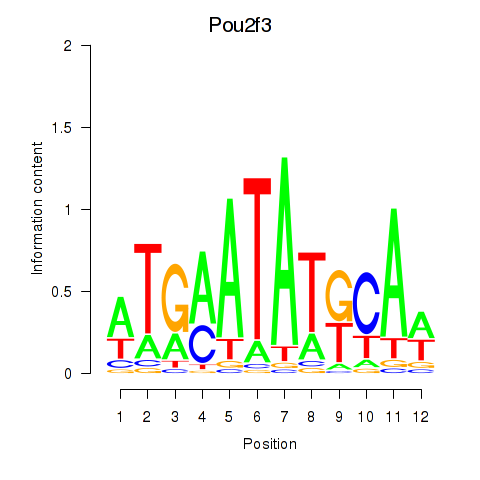
Transcription factors associated with Pou2f3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pou2f3 | ENSMUSG00000032015.9 | Pou2f3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.8 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 1.1 | 3.2 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.0 | 3.1 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.6 | 1.9 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.6 | 3.7 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.6 | 3.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 | 5.9 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.4 | 1.3 | GO:0002725 | negative regulation of antigen processing and presentation(GO:0002578) negative regulation of T cell cytokine production(GO:0002725) |
| 0.4 | 1.7 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.4 | 1.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.3 | 2.3 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.3 | 1.9 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.3 | 2.1 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.3 | 1.2 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.3 | 1.7 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 1.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 2.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.3 | 1.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 1.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.3 | 0.8 | GO:0030210 | heparin biosynthetic process(GO:0030210) Tie signaling pathway(GO:0048014) |
| 0.2 | 1.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 1.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 0.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 0.7 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.2 | 1.3 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.2 | 1.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 0.6 | GO:0042737 | drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.2 | 0.6 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.2 | 0.5 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.2 | 1.5 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 1.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.0 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 0.6 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.8 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 1.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.4 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.1 | 0.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.3 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 1.8 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 | 1.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 1.5 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 2.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.2 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.1 | 1.5 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.5 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.1 | 0.5 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 1.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 1.0 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.3 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.6 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.4 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 2.2 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.9 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 1.1 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 4.8 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.8 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 4.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.0 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.7 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.4 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 2.4 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) inositol lipid-mediated signaling(GO:0048017) |
| 0.0 | 0.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.2 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 1.1 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:1901724 | positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 1.2 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 1.0 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 1.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.2 | GO:0048754 | branching morphogenesis of an epithelial tube(GO:0048754) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.5 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.4 | 1.9 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.4 | 1.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 1.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 1.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 1.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 1.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 0.6 | GO:0071920 | cleavage body(GO:0071920) |
| 0.2 | 1.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 0.6 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 1.5 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.9 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 2.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.6 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 1.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 0.2 | GO:0005940 | septin ring(GO:0005940) |
| 0.1 | 5.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 4.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.2 | GO:1990423 | Dsl1p complex(GO:0070939) RZZ complex(GO:1990423) |
| 0.1 | 2.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 1.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 3.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.9 | 2.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.8 | 5.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.8 | 3.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.6 | 1.9 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.6 | 1.8 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.4 | 4.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.3 | 1.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.3 | 1.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 1.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 1.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 1.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 1.9 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.2 | 1.7 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.2 | 0.5 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 0.6 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.1 | 1.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 2.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 1.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 1.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.3 | GO:1990459 | beta-2-microglobulin binding(GO:0030881) transferrin receptor binding(GO:1990459) |
| 0.1 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 2.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.2 | GO:0016160 | alpha-amylase activity(GO:0004556) amylase activity(GO:0016160) |
| 0.1 | 0.6 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 1.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.0 | 1.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 3.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.6 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 1.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 4.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.2 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.2 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 5.8 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 6.7 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.1 | 2.0 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.8 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.1 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.3 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | PID_IL27_PATHWAY | IL27-mediated signaling events |
| 0.0 | 5.2 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.0 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.5 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.5 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 1.5 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.8 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.3 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.0 | 2.2 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID_E2F_PATHWAY | E2F transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.2 | 2.3 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 2.5 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 2.7 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.8 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.3 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.6 | REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.2 | REACTOME_NEGATIVE_REGULATION_OF_THE_PI3K_AKT_NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.3 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.6 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.1 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.6 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.7 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.1 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.5 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.8 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME_MRNA_CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.8 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 3.1 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.1 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.6 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.3 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.7 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 4.7 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME_DNA_REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.3 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |