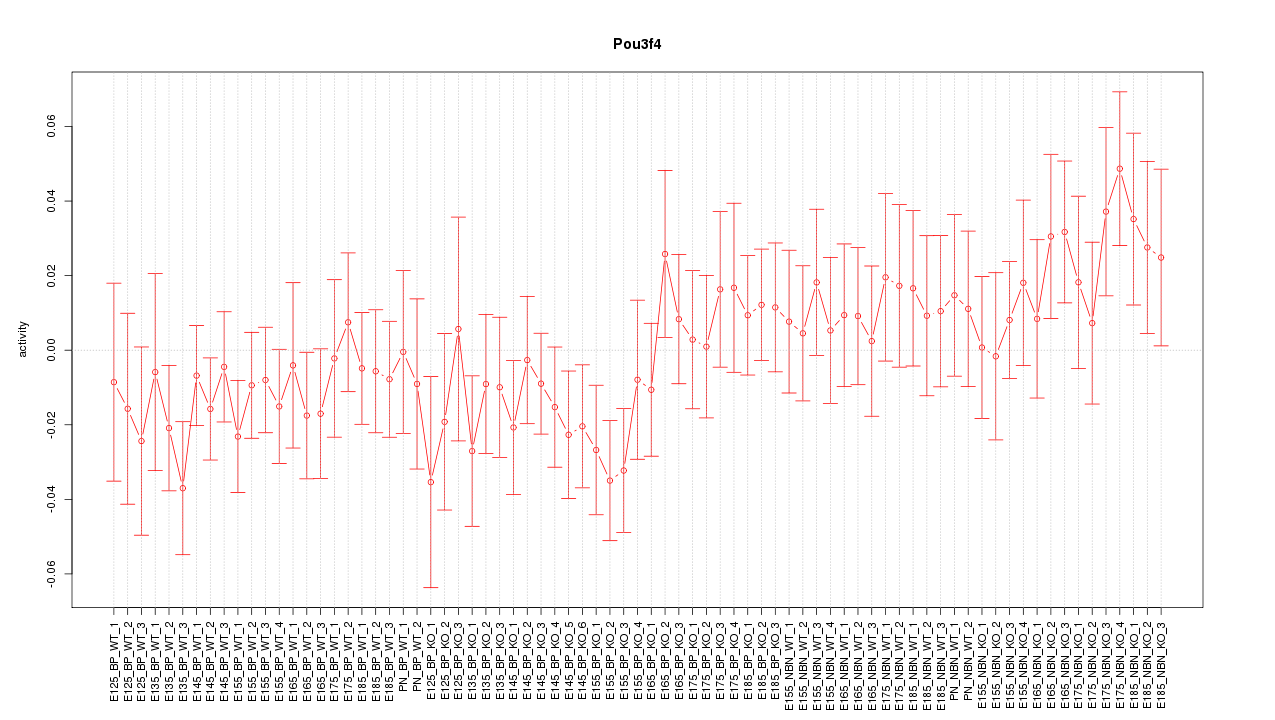
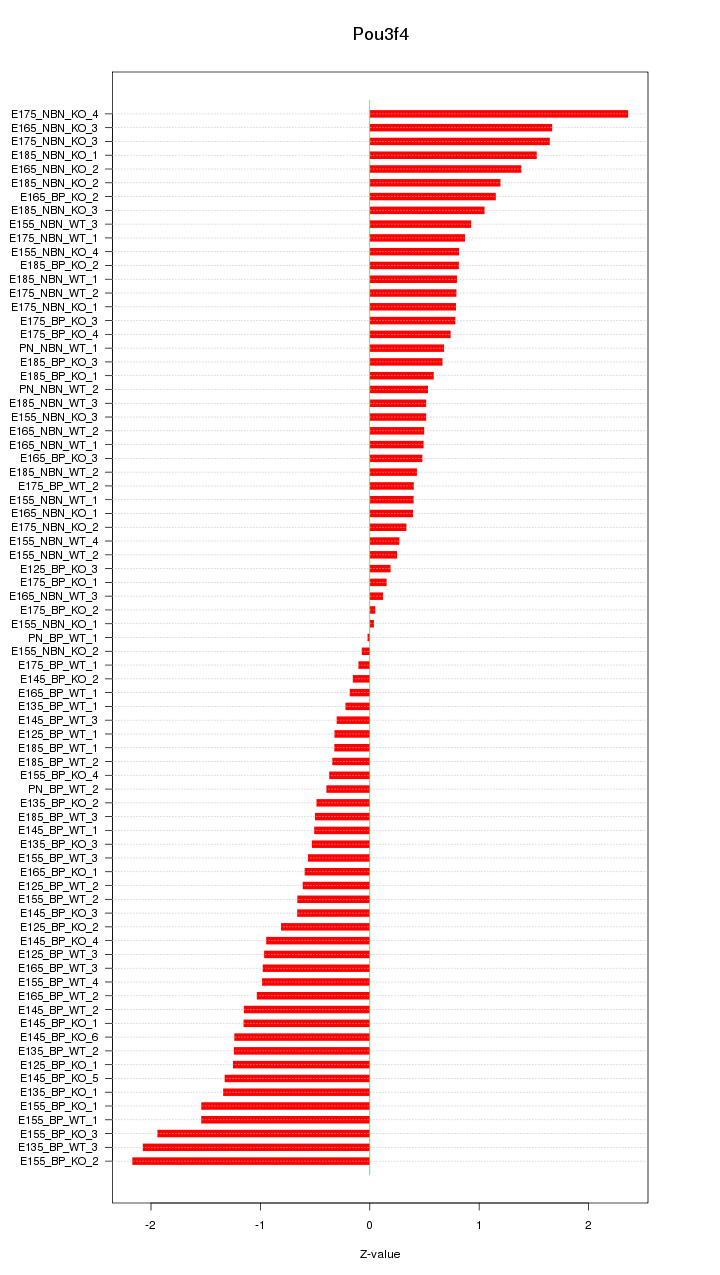
Motif ID: Pou3f4
Z-value: 0.930
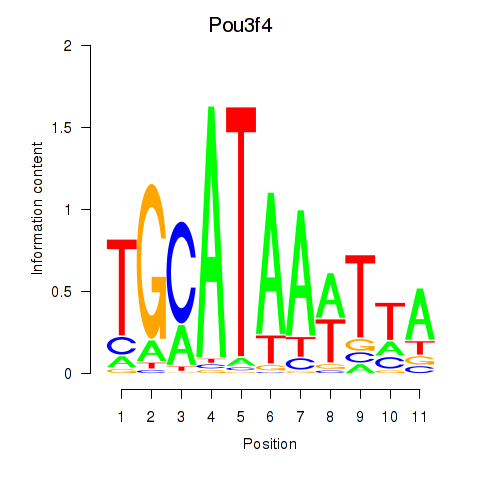
Transcription factors associated with Pou3f4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Pou3f4 | ENSMUSG00000056854.3 | Pou3f4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f4 | mm10_v2_chrX_+_110814390_110814413 | -0.35 | 2.1e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 32.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 3.4 | 13.4 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 3.0 | 9.0 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 2.7 | 8.0 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 2.4 | 16.6 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.8 | 7.3 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 1.3 | 5.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.3 | 3.9 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) |
| 1.0 | 4.1 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.9 | 2.7 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.8 | 2.5 | GO:1900141 | regulation of oligodendrocyte apoptotic process(GO:1900141) negative regulation of oligodendrocyte apoptotic process(GO:1900142) |
| 0.8 | 3.2 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.8 | 12.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.8 | 5.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.5 | 6.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.5 | 1.5 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.4 | 1.3 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.4 | 5.8 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.4 | 3.8 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.4 | 6.8 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.4 | 2.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 8.8 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.3 | 1.6 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.3 | 2.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 3.6 | GO:0034312 | diol biosynthetic process(GO:0034312) |
| 0.3 | 1.4 | GO:0051940 | regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.3 | 5.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.3 | 3.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.3 | 1.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 4.3 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.2 | 1.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 1.0 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 1.0 | GO:1901526 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of macromitophagy(GO:1901526) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.2 | 5.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.2 | 1.7 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.2 | 7.3 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.2 | 0.6 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.2 | 3.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 2.0 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 0.7 | GO:0061743 | motor learning(GO:0061743) |
| 0.2 | 2.5 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 1.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.8 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 1.0 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.1 | 9.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.1 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 3.6 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 0.8 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 3.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.6 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.5 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 1.0 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 2.5 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.4 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 2.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.3 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 1.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 2.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 4.2 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 5.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.4 | GO:0072534 | perineuronal net(GO:0072534) |
| 1.2 | 12.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 4.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.7 | 10.0 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.6 | 2.5 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.5 | 5.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 2.7 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 1.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 17.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 1.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 0.6 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 0.8 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 1.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 5.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.0 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 3.7 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 8.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 11.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 0.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.8 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 3.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 3.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 12.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 7.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 7.3 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 3.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 3.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.7 | GO:0001650 | fibrillar center(GO:0001650) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 16.6 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 2.7 | 8.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 2.3 | 9.0 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) A2A adenosine receptor binding(GO:0031687) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 1.8 | 7.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 1.4 | 4.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 1.0 | 5.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.8 | 13.4 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.7 | 3.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.7 | 5.3 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.6 | 4.3 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.6 | 3.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.5 | 3.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.5 | 2.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 7.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.4 | 2.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.4 | 2.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.4 | 3.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.3 | 1.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 1.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.3 | 3.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 1.0 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.2 | 0.8 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.1 | 3.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 4.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 5.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 1.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.3 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 5.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 2.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 2.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.0 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 2.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 5.5 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 5.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 6.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 3.8 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 5.1 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 1.0 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 29.3 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 0.4 | 8.0 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 16.6 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.4 | 8.8 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.2 | 3.1 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.2 | 4.1 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 9.5 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.1 | 16.0 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.5 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 2.7 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.0 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.7 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | NABA_COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 16.6 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.4 | 4.1 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 8.8 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 9.1 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 3.0 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 4.3 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.1 | 5.2 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.1 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 0.8 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 2.7 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.2 | REACTOME_REGULATION_OF_GLUCOKINASE_BY_GLUCOKINASE_REGULATORY_PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 2.1 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 3.7 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.1 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.6 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.8 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME_ACTIVATION_OF_THE_MRNA_UPON_BINDING_OF_THE_CAP_BINDING_COMPLEX_AND_EIFS_AND_SUBSEQUENT_BINDING_TO_43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.0 | 0.4 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |