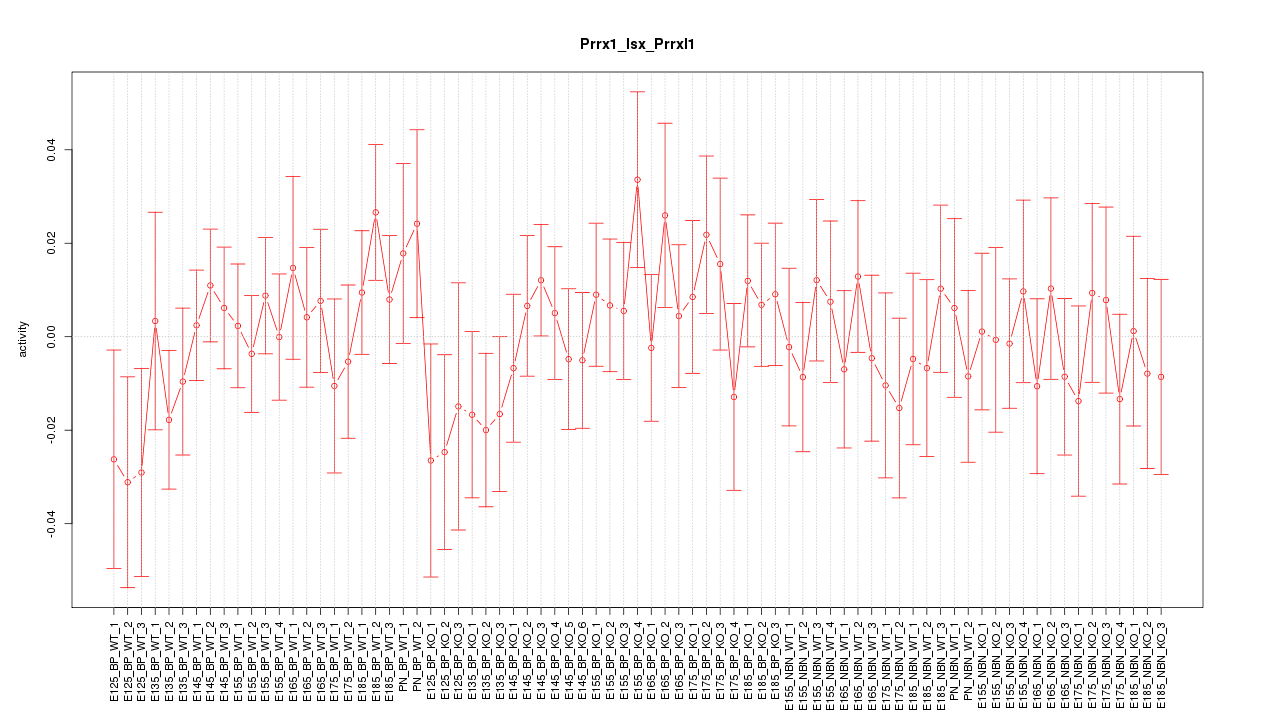
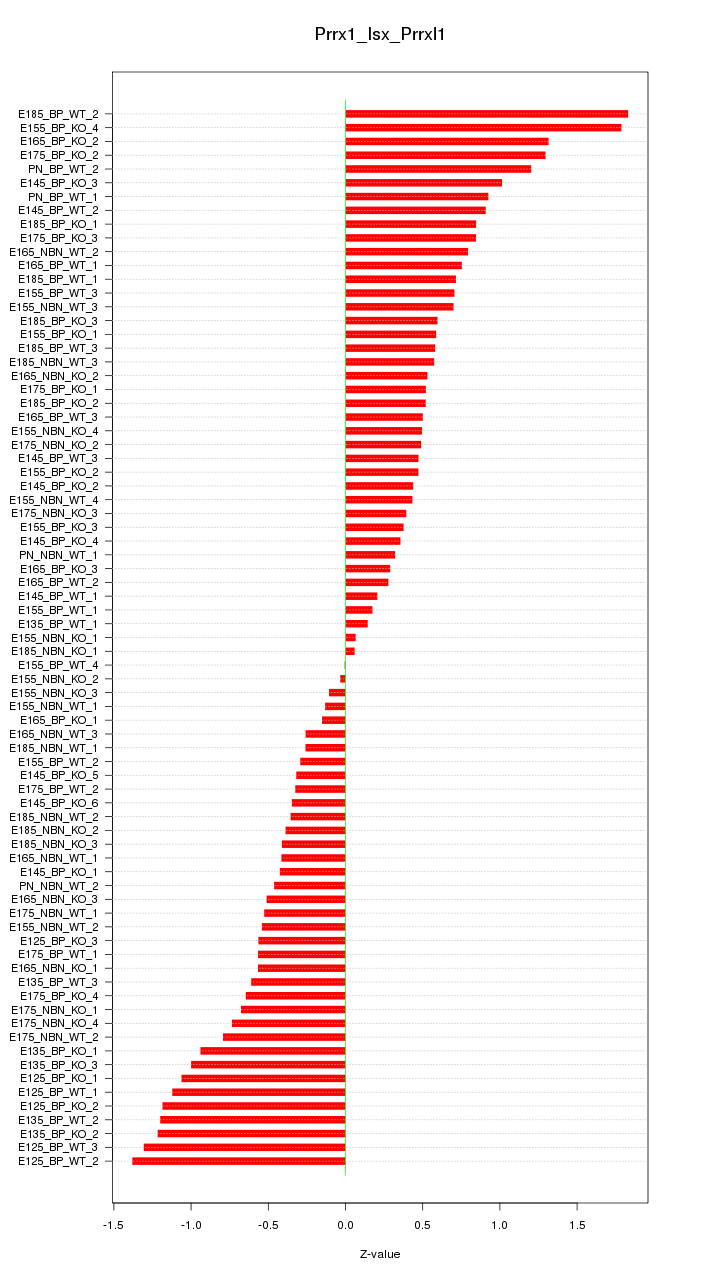
Motif ID: Prrx1_Isx_Prrxl1
Z-value: 0.728

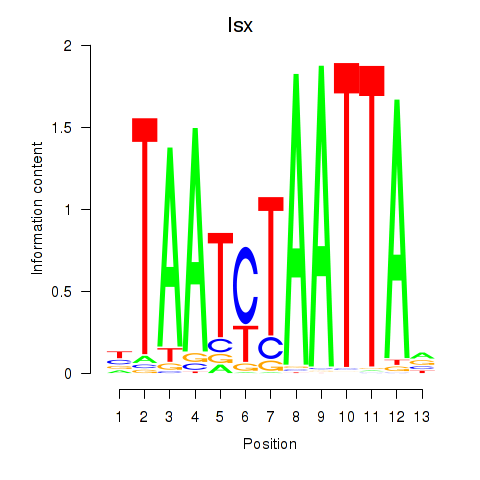
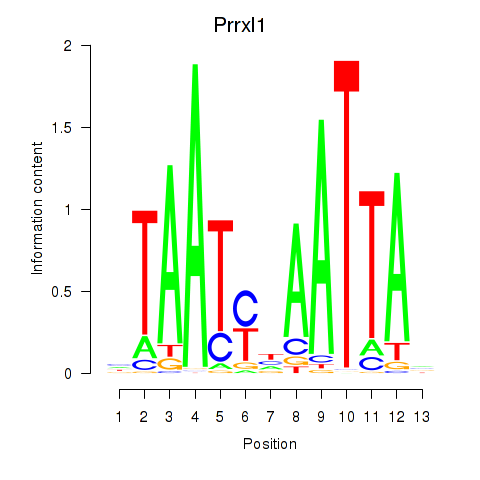
Transcription factors associated with Prrx1_Isx_Prrxl1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Isx | ENSMUSG00000031621.3 | Isx |
| Prrx1 | ENSMUSG00000026586.10 | Prrx1 |
| Prrxl1 | ENSMUSG00000041730.7 | Prrxl1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prrxl1 | mm10_v2_chr14_+_32599922_32599932 | -0.35 | 1.8e-03 | Click! |
| Prrx1 | mm10_v2_chr1_-_163313661_163313710 | 0.33 | 3.9e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.3 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.8 | 7.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.1 | 3.3 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 1.1 | 3.2 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.8 | 3.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 | 5.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.6 | 2.9 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.6 | 9.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.5 | 1.6 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.5 | 1.4 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.5 | 1.4 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.4 | 1.3 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.4 | 1.3 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.4 | 1.3 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.4 | 2.9 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.4 | 8.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.4 | 1.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.3 | 2.0 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.3 | 1.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.3 | 1.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.3 | 1.5 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.3 | 1.5 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.3 | 1.5 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.3 | 0.8 | GO:2000564 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.3 | 1.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.3 | 1.3 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.3 | 0.5 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.3 | 1.0 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 1.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 0.7 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.2 | 1.0 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.2 | 0.6 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 0.4 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.2 | 4.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 0.6 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.2 | 1.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 | 0.5 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 4.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.2 | 2.4 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.2 | 1.7 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.2 | 3.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.8 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.2 | 0.8 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 0.9 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.2 | 0.6 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 0.8 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.2 | 0.6 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.2 | 1.1 | GO:0060315 | response to redox state(GO:0051775) negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.2 | 0.9 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.2 | 1.7 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 1.0 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.7 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.1 | 1.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 11.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 5.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.6 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.1 | 0.4 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.1 | 0.7 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 1.1 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 0.3 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.3 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.5 | GO:1990564 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 4.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 0.3 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.1 | 2.6 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 1.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.7 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.1 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.3 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.9 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.3 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.4 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 1.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.1 | 2.0 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.2 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 1.0 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 0.9 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 1.7 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.3 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.3 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.3 | GO:0098909 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.5 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.0 | 0.1 | GO:1903546 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 1.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 1.6 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.3 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 2.7 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 2.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 1.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 4.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.3 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 1.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.2 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.0 | 1.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 4.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.3 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 2.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.8 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:1901663 | quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.1 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.0 | GO:0019085 | early viral transcription(GO:0019085) late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 | 0.6 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.5 | 6.5 | GO:0042581 | specific granule(GO:0042581) |
| 0.3 | 1.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.3 | 1.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 1.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.3 | 1.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.2 | 1.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 2.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 3.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 7.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 5.7 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 0.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.6 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 2.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.3 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 4.6 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 0.9 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.5 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 4.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 3.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 4.3 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 1.2 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 7.6 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 8.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 4.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.8 | GO:0032993 | protein-DNA complex(GO:0032993) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 11.8 | GO:0019841 | retinol binding(GO:0019841) |
| 1.2 | 7.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.9 | 2.6 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.8 | 3.3 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.7 | 3.6 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.4 | 1.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.4 | 1.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.4 | 1.7 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.4 | 1.3 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.4 | 5.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.4 | 2.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.4 | 1.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.3 | 1.0 | GO:0004980 | melanocortin receptor activity(GO:0004977) melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.3 | 1.3 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.3 | 3.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.3 | 1.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.3 | 1.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 1.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 1.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 1.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 1.8 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.2 | 1.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 2.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 3.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 0.6 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.2 | 0.5 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 4.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 2.0 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 9.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 4.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.9 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 1.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 3.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 5.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 3.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.5 | GO:0097493 | muscle alpha-actinin binding(GO:0051371) structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.6 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled adenosine receptor activity(GO:0001609) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.4 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 1.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 3.0 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.3 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 0.7 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 1.0 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 1.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 1.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.5 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 3.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 4.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 6.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.0 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 1.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.8 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.1 | 5.5 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 4.4 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.7 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.2 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.8 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.1 | 3.3 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 8.5 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.3 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 2.9 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.0 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 3.4 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.5 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.1 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 5.9 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.4 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.2 | PID_P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.1 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.2 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 1.4 | REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 1.8 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 7.1 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 2.2 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.1 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.8 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.4 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.1 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 5.8 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 4.8 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.7 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.5 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 3.2 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.7 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.0 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.0 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.1 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.4 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 3.1 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 5.0 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 2.6 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.9 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.0 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.3 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.5 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.9 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.0 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.5 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 2.9 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.5 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 3.7 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.6 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.5 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |