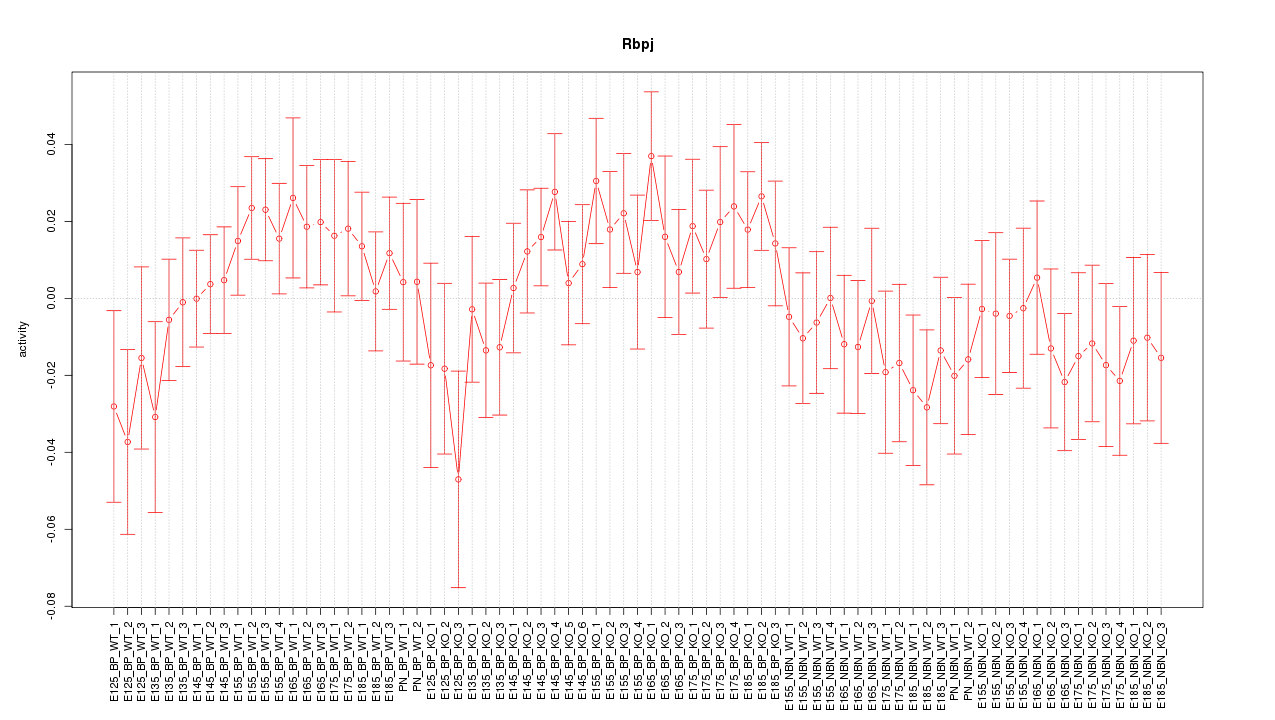
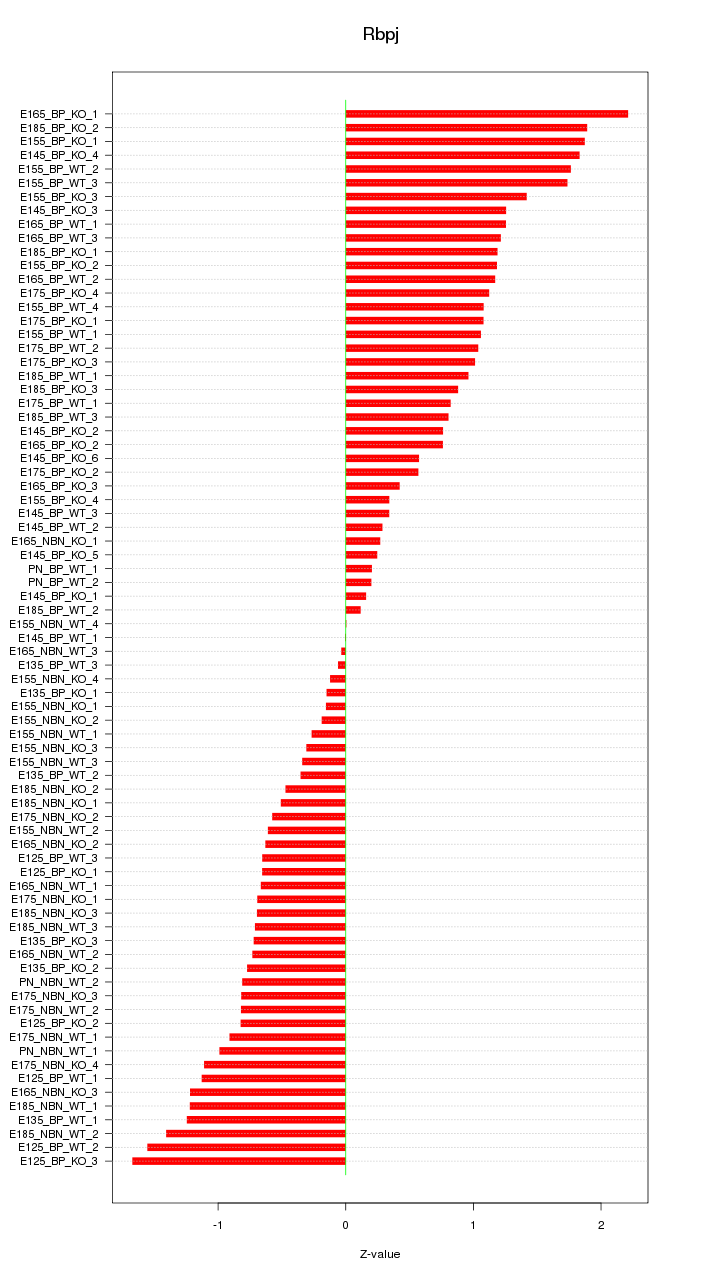
Motif ID: Rbpj
Z-value: 0.953
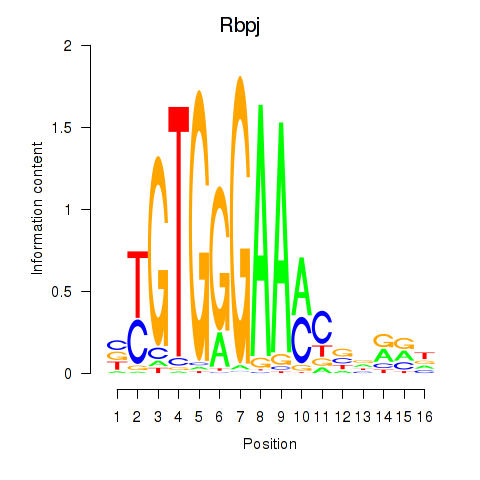
Transcription factors associated with Rbpj:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rbpj | ENSMUSG00000039191.6 | Rbpj |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rbpj | mm10_v2_chr5_+_53590453_53590574 | 0.51 | 1.7e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 15.7 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 2.9 | 8.8 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 2.2 | 4.4 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 1.7 | 5.1 | GO:0003195 | tricuspid valve formation(GO:0003195) |
| 1.6 | 4.7 | GO:0060129 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 1.3 | 5.4 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.9 | 6.4 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.8 | 7.0 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.7 | 2.8 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.7 | 2.7 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) |
| 0.6 | 4.9 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.6 | 10.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.5 | 1.6 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.5 | 4.7 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.5 | 1.9 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.5 | 1.4 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.4 | 1.2 | GO:0032240 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.4 | 2.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.4 | 1.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.3 | 1.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.3 | 1.4 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.3 | 1.3 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.3 | 1.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.3 | 2.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.3 | 1.6 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.3 | 2.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 3.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 | 3.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 0.7 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.2 | 1.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 1.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 0.2 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.2 | 1.5 | GO:0098707 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.2 | 1.8 | GO:0046959 | habituation(GO:0046959) |
| 0.2 | 3.9 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.2 | 4.4 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.2 | 0.5 | GO:0002489 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.2 | 2.0 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.2 | 2.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 4.5 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 1.0 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.7 | GO:0015871 | astrocyte activation involved in immune response(GO:0002265) choline transport(GO:0015871) |
| 0.1 | 0.8 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 1.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 2.2 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.1 | 2.0 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.8 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 1.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 2.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 1.8 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 | 2.7 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 1.0 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 1.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 2.4 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.1 | 1.4 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 1.3 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 5.9 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.1 | 0.8 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.4 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.1 | 0.3 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) DNA endoreduplication(GO:0042023) |
| 0.1 | 2.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.5 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 0.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 1.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:1904398 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.2 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 2.7 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 1.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.8 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.7 | 4.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.7 | 2.8 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.7 | 2.7 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.5 | 2.0 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.5 | 10.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.5 | 6.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 1.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 2.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 1.5 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 2.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 1.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.2 | 2.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 1.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.8 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.5 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 4.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 7.0 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 2.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 15.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.6 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 2.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.8 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 4.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.9 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 11.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.6 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 3.7 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 4.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 7.0 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 1.7 | 5.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.0 | 15.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.7 | 2.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.7 | 5.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.7 | 4.7 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.6 | 1.9 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) C-X-C chemokine binding(GO:0019958) |
| 0.5 | 3.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.5 | 2.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.5 | 2.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.4 | 1.3 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.4 | 2.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.4 | 2.8 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.3 | 1.6 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.3 | 1.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 3.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 1.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.2 | 1.5 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 1.0 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 3.9 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 1.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 1.0 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.6 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 0.8 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 3.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.5 | GO:0032450 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 0.5 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 4.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.4 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.3 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 0.5 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 3.8 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.1 | 2.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.3 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.7 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 4.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.7 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.7 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 2.0 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 9.2 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 8.8 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.6 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 1.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) dynein complex binding(GO:0070840) |
| 0.0 | 2.6 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 1.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 5.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.8 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 1.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 3.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.5 | GO:0001076 | transcription factor activity, RNA polymerase II transcription factor binding(GO:0001076) |
| 0.0 | 1.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 12.4 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.8 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 13.3 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 7.0 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 16.6 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.1 | 4.4 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 4.7 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 5.6 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 1.9 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 0.9 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.4 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.4 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.4 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.0 | 1.3 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID_ATR_PATHWAY | ATR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 15.7 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.4 | 7.0 | REACTOME_PLATELET_SENSITIZATION_BY_LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 6.9 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 3.9 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 2.7 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 4.7 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.2 | 2.1 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.1 | 4.5 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.3 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.4 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 6.2 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 3.2 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.0 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.9 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 4.6 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.9 | REACTOME_REGULATED_PROTEOLYSIS_OF_P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 1.8 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.3 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 1.2 | REACTOME_NEP_NS2_INTERACTS_WITH_THE_CELLULAR_EXPORT_MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.6 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 2.4 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.8 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 1.0 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.7 | REACTOME_INFLUENZA_VIRAL_RNA_TRANSCRIPTION_AND_REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 1.4 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.1 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.4 | REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |