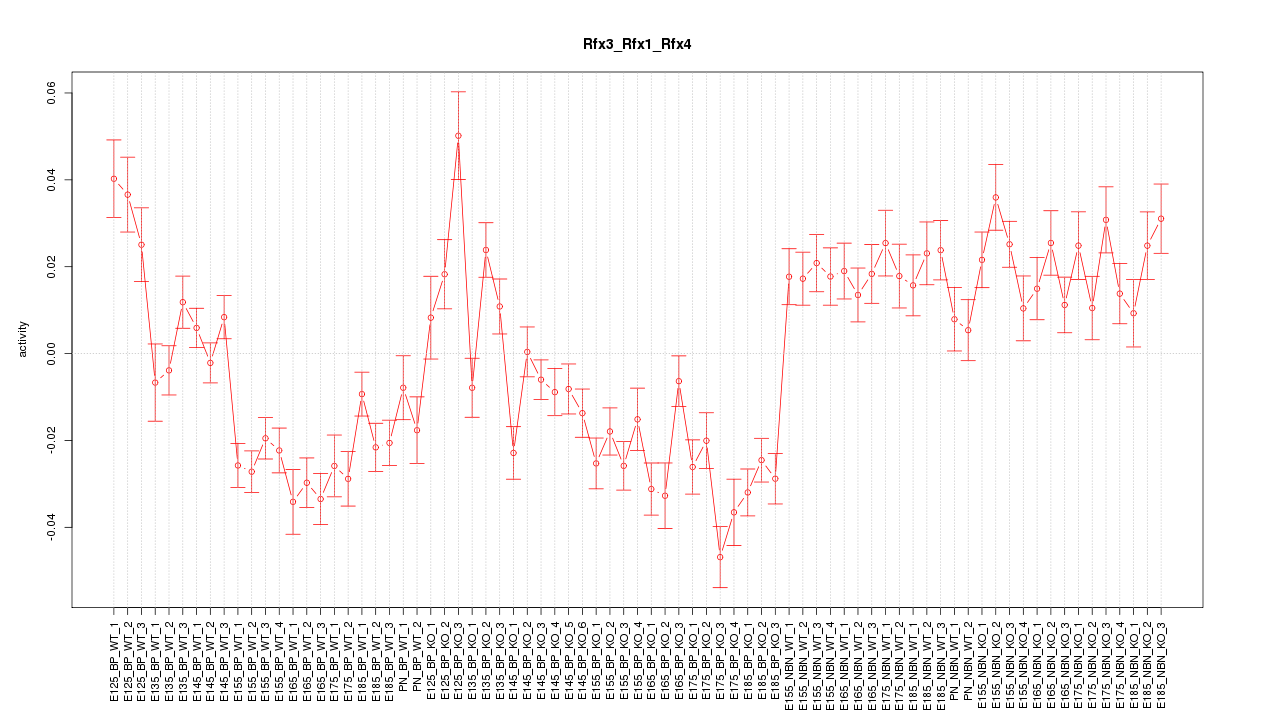
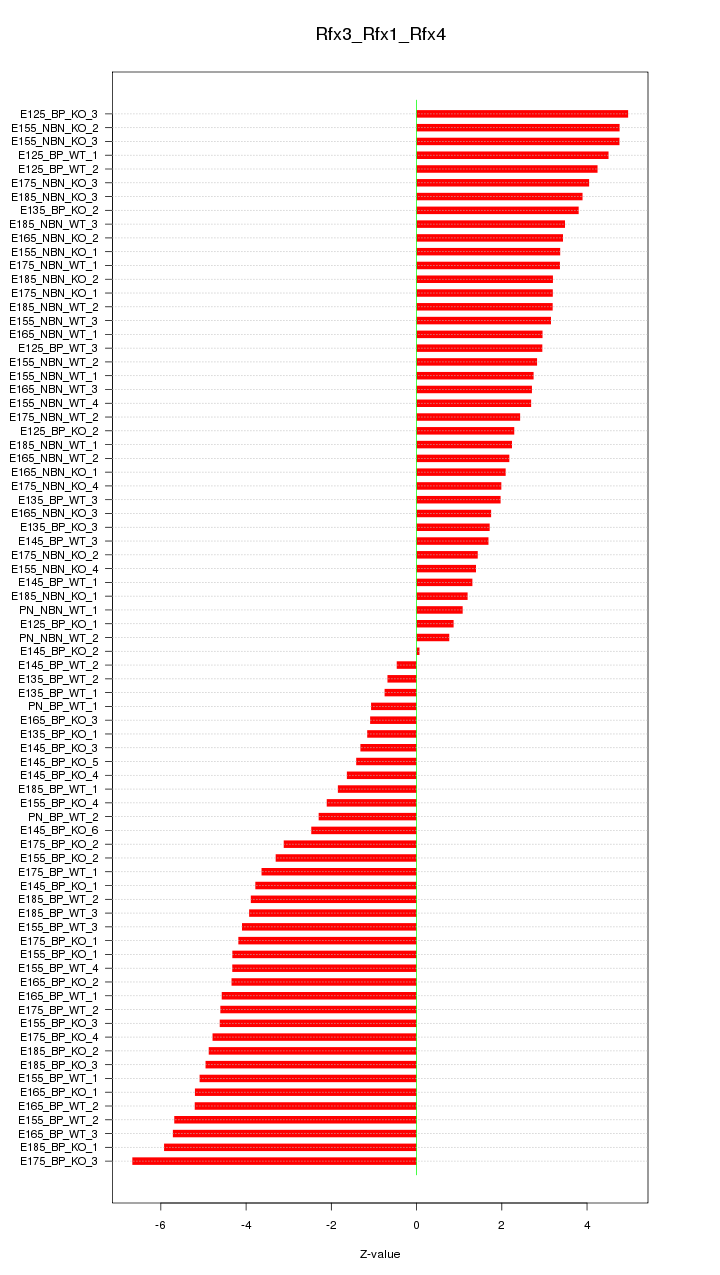
Motif ID: Rfx3_Rfx1_Rfx4
Z-value: 3.419
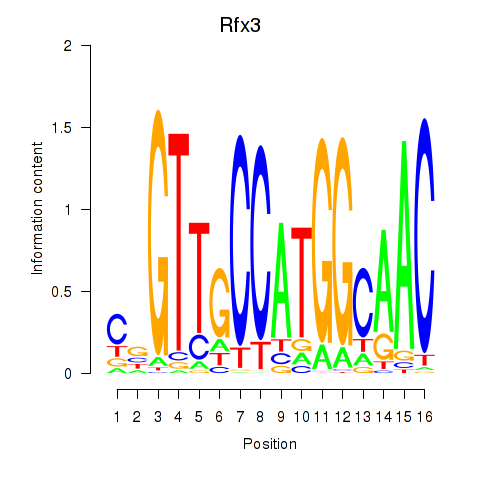
Transcription factors associated with Rfx3_Rfx1_Rfx4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rfx1 | ENSMUSG00000031706.6 | Rfx1 |
| Rfx3 | ENSMUSG00000040929.10 | Rfx3 |
| Rfx4 | ENSMUSG00000020037.9 | Rfx4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx4 | mm10_v2_chr10_+_84838143_84838153 | -0.22 | 5.6e-02 | Click! |
| Rfx1 | mm10_v2_chr8_+_84066824_84066882 | -0.20 | 7.6e-02 | Click! |
| Rfx3 | mm10_v2_chr19_-_28010995_28011054 | -0.12 | 3.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.9 | 167.3 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 6.8 | 27.3 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 6.6 | 33.0 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 6.0 | 30.2 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 5.0 | 14.9 | GO:1990523 | negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) bone regeneration(GO:1990523) |
| 4.3 | 12.9 | GO:0009826 | unidimensional cell growth(GO:0009826) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 4.3 | 8.6 | GO:0021508 | ventral midline development(GO:0007418) floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 3.9 | 27.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 3.8 | 11.3 | GO:2000836 | negative regulation of glucagon secretion(GO:0070093) positive regulation of androgen secretion(GO:2000836) positive regulation of testosterone secretion(GO:2000845) |
| 3.1 | 3.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 3.1 | 12.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 2.9 | 11.6 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 2.7 | 5.4 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 2.7 | 16.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 2.6 | 10.4 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 2.6 | 10.3 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 2.6 | 23.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 2.5 | 7.6 | GO:0097402 | neuroblast migration(GO:0097402) |
| 2.5 | 70.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 2.2 | 8.8 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 2.1 | 8.3 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 2.0 | 18.4 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 1.9 | 5.6 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 1.8 | 3.7 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 1.8 | 14.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 1.8 | 5.3 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 1.7 | 33.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 1.7 | 6.8 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 1.6 | 31.0 | GO:0071625 | vocalization behavior(GO:0071625) |
| 1.6 | 8.0 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 1.6 | 9.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.5 | 7.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 1.5 | 14.8 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 1.3 | 4.0 | GO:0016598 | protein arginylation(GO:0016598) |
| 1.3 | 30.6 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 1.3 | 3.9 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 1.3 | 5.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 1.3 | 3.8 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 1.2 | 7.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 1.2 | 5.9 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 1.2 | 25.8 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 1.2 | 8.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 1.1 | 38.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 1.1 | 4.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.1 | 5.3 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 1.1 | 7.4 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 1.0 | 3.1 | GO:0015886 | heme transport(GO:0015886) |
| 1.0 | 6.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 1.0 | 6.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 1.0 | 4.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 1.0 | 2.9 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.9 | 7.6 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.9 | 2.8 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.9 | 22.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.9 | 73.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.8 | 8.4 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.8 | 0.8 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 0.8 | 13.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.8 | 2.4 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.8 | 3.2 | GO:0086042 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.8 | 9.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.8 | 10.0 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.8 | 2.3 | GO:0099541 | diacylglycerol catabolic process(GO:0046340) retrograde trans-synaptic signaling(GO:0098917) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.8 | 7.5 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.7 | 8.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.7 | 4.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.7 | 10.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.7 | 2.1 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.7 | 23.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.7 | 4.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.7 | 6.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.7 | 11.3 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.7 | 53.6 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.7 | 2.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.7 | 2.6 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.6 | 2.6 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.6 | 22.2 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.6 | 3.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.6 | 5.0 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.6 | 1.8 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.6 | 2.4 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.6 | 1.7 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.6 | 5.1 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.5 | 6.5 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.5 | 17.0 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.5 | 2.6 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.5 | 7.8 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.5 | 1.5 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.5 | 1.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.5 | 2.0 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.5 | 1.4 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.5 | 40.3 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.5 | 7.7 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.5 | 1.4 | GO:1902220 | positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.5 | 6.3 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.4 | 2.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.4 | 1.3 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.4 | 2.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.4 | 1.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.4 | 3.3 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.4 | 1.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) formin-nucleated actin cable assembly(GO:0070649) |
| 0.4 | 22.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.4 | 7.3 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.4 | 4.8 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.4 | 21.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.4 | 4.7 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.4 | 3.5 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.4 | 4.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.4 | 1.1 | GO:0003356 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.4 | 1.9 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.4 | 4.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.4 | 12.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.4 | 2.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.4 | 8.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.3 | 9.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.3 | 1.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 4.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 4.4 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.3 | 5.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.3 | 2.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 4.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.3 | 1.7 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.3 | 1.8 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.3 | 8.8 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.3 | 15.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.3 | 0.9 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.3 | 2.0 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.3 | 1.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.3 | 2.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.3 | 1.9 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.3 | 10.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 3.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 11.0 | GO:0034766 | negative regulation of ion transmembrane transport(GO:0034766) |
| 0.2 | 3.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 0.7 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.2 | 3.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.2 | 2.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 1.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 2.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.2 | 0.9 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.2 | 2.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 12.2 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.2 | 4.4 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.2 | 0.8 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 1.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 4.4 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.2 | 1.2 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.2 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 2.8 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 5.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 5.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.2 | 6.6 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) |
| 0.2 | 3.3 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.2 | 2.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 0.9 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.2 | 2.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 2.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 | 2.1 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.2 | 0.7 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 1.2 | GO:0061092 | involuntary skeletal muscle contraction(GO:0003011) regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 | 7.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.2 | 10.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.2 | 1.8 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.2 | 4.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.2 | 0.5 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.2 | 0.6 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.5 | GO:0032439 | endosome localization(GO:0032439) |
| 0.1 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 1.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.8 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.1 | 1.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.9 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 6.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.3 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 1.0 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 3.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 2.0 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.1 | 1.4 | GO:0097154 | GABAergic neuron differentiation(GO:0097154) |
| 0.1 | 1.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 3.5 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.1 | 1.4 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 2.5 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 1.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 | 3.3 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.9 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 1.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 16.0 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 2.1 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 2.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 9.1 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 3.0 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) negative regulation of actin nucleation(GO:0051126) |
| 0.1 | 2.0 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 0.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 8.5 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 2.4 | GO:0061371 | determination of heart left/right asymmetry(GO:0061371) |
| 0.1 | 0.2 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 0.6 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 3.4 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.1 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 2.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 6.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 1.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 2.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 4.3 | GO:0060563 | neuroepithelial cell differentiation(GO:0060563) |
| 0.1 | 0.3 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.9 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 3.1 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.1 | 0.9 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.3 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 1.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.4 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.0 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0032927 | regulation of mRNA export from nucleus(GO:0010793) positive regulation of activin receptor signaling pathway(GO:0032927) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 | 1.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 4.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.5 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 6.6 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.5 | GO:0050965 | detection of temperature stimulus(GO:0016048) detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 6.7 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 12.9 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.5 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) |
| 0.0 | 0.1 | GO:0006684 | sphingomyelin metabolic process(GO:0006684) |
| 0.0 | 2.5 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 5.0 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 1.0 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.6 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 1.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.8 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) ribonucleoprotein complex export from nucleus(GO:0071426) |
| 0.0 | 1.8 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 2.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.3 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.4 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 1.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.2 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
| 0.0 | 0.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 2.1 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 2.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.4 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.1 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 52.0 | 156.0 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 10.0 | 39.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 2.7 | 10.6 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 1.8 | 16.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.8 | 5.4 | GO:0016939 | kinesin II complex(GO:0016939) |
| 1.7 | 8.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.5 | 16.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 1.3 | 8.0 | GO:0000235 | astral microtubule(GO:0000235) |
| 1.2 | 9.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 1.0 | 16.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 1.0 | 12.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 1.0 | 23.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.9 | 19.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.9 | 3.8 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.9 | 9.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.8 | 6.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.8 | 10.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.8 | 11.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.8 | 2.4 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.7 | 83.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.7 | 10.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.7 | 19.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.7 | 10.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.7 | 5.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.7 | 20.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.7 | 18.4 | GO:0031430 | M band(GO:0031430) |
| 0.7 | 11.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.6 | 21.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.6 | 6.4 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.6 | 14.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.6 | 6.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.5 | 3.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.5 | 7.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.5 | 11.3 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.5 | 1.9 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.5 | 2.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.5 | 8.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.5 | 19.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.4 | 8.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 1.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.4 | 8.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.4 | 2.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 3.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.4 | 1.9 | GO:1990745 | EARP complex(GO:1990745) |
| 0.3 | 8.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.3 | 12.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 4.2 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.3 | 1.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 2.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 4.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.3 | 1.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.3 | 3.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 1.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.3 | 9.8 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) |
| 0.3 | 65.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.3 | 24.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 10.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 1.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.3 | 3.5 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.2 | 0.5 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.2 | 2.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 2.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 2.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 34.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.2 | 4.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 1.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 2.9 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 7.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 3.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 7.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 2.4 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.2 | 4.4 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 8.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.2 | 2.3 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.2 | 3.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 11.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 7.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 2.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 0.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 7.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 9.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.0 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.6 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 4.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 5.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 5.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 23.7 | GO:0019867 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.1 | 5.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 43.2 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 0.4 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 4.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 11.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 3.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 5.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 10.4 | GO:0031514 | motile cilium(GO:0031514) |
| 0.1 | 3.4 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 3.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 6.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.0 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.3 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 1.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.6 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 2.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 1.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 20.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 2.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 43.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 1.2 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 8.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 7.2 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 3.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 70.1 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.8 | 50.3 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 15.8 | 189.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 8.1 | 64.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 6.9 | 27.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 2.9 | 11.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 2.4 | 9.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 2.1 | 16.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.9 | 9.6 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 1.8 | 5.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 1.6 | 9.8 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.4 | 11.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 1.4 | 5.5 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 1.3 | 7.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.3 | 3.8 | GO:0005119 | smoothened binding(GO:0005119) |
| 1.3 | 3.8 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 1.2 | 3.5 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 1.1 | 5.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.1 | 3.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 1.1 | 4.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.0 | 3.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.0 | 30.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 1.0 | 4.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 1.0 | 4.8 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.9 | 27.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.9 | 41.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.9 | 7.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.9 | 2.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.8 | 3.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.8 | 4.8 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.8 | 3.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.8 | 22.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.7 | 25.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.7 | 32.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.7 | 9.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.7 | 7.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.7 | 10.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.7 | 11.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.7 | 8.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.6 | 1.9 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.6 | 3.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.6 | 7.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.6 | 7.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.6 | 4.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.6 | 7.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.6 | 3.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.6 | 25.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.6 | 21.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.5 | 1.6 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.5 | 10.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.5 | 5.3 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.5 | 24.2 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.5 | 1.4 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.5 | 3.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.5 | 7.6 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.5 | 1.4 | GO:0070736 | protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) |
| 0.5 | 11.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.4 | 2.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.4 | 1.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.4 | 1.8 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.4 | 1.2 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.4 | 6.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.4 | 5.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 2.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.4 | 4.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.4 | 2.0 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.4 | 1.9 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.4 | 31.0 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.4 | 2.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.4 | 2.7 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.4 | 12.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 7.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 3.4 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.3 | 18.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 2.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.3 | 3.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 8.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 2.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 3.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.3 | 8.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 0.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 5.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 4.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.3 | 4.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 3.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 8.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 4.9 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 64.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.2 | 1.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 1.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 0.7 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 1.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 0.5 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.2 | 7.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 1.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.2 | 3.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 17.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 23.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.2 | 7.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 2.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 2.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 1.0 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 5.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.2 | 4.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 6.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 7.8 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.2 | 9.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 17.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 10.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 2.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 2.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 3.0 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.2 | 5.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 4.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 2.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.2 | 1.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 4.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 5.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 5.7 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 2.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 6.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 13.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 3.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 2.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 10.3 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.5 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 2.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 21.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 6.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 12.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 6.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 34.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 1.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 1.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 1.8 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.5 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 6.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.5 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.1 | 0.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 3.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 2.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.5 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 3.0 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 1.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 3.0 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.1 | 2.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 3.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 5.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 2.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.6 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 10.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 5.3 | GO:0016874 | ligase activity(GO:0016874) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 2.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.7 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 1.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.1 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 1.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 156.0 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.0 | 33.0 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.5 | 16.7 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.5 | 3.7 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.4 | 14.8 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.4 | 9.4 | ST_GRANULE_CELL_SURVIVAL_PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.4 | 38.5 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.3 | 3.3 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.3 | 16.5 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.3 | 4.8 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.2 | 6.5 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.2 | 19.3 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.2 | 4.8 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.2 | 25.2 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.2 | 10.3 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.2 | 5.4 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.2 | 6.4 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.2 | 1.6 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 1.1 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 6.9 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.2 | 1.6 | PID_AVB3_OPN_PATHWAY | Osteopontin-mediated events |
| 0.2 | 1.4 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 18.3 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 7.5 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 8.6 | PID_P73PATHWAY | p73 transcription factor network |
| 0.1 | 3.7 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.1 | 6.1 | PID_CERAMIDE_PATHWAY | Ceramide signaling pathway |
| 0.1 | 8.3 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.6 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.1 | 5.2 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.9 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 14.2 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 3.1 | PID_LYSOPHOSPHOLIPID_PATHWAY | LPA receptor mediated events |
| 0.1 | 2.1 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.2 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.5 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.3 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.6 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 5.8 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.3 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 161.5 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 2.9 | 78.0 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 2.0 | 49.2 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 1.2 | 30.6 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 1.0 | 25.0 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.9 | 12.1 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.8 | 3.3 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.8 | 10.4 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.8 | 24.8 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.7 | 12.3 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.7 | 11.4 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.5 | 6.5 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.5 | 45.0 | REACTOME_REGULATION_OF_INSULIN_SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.5 | 7.7 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.5 | 8.4 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 4.9 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.4 | 18.0 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.4 | 7.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.4 | 31.6 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.4 | 12.9 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 5.8 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.3 | 10.3 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.3 | 11.1 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.3 | 5.4 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 12.5 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.3 | 5.6 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 1.2 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 7.3 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 13.8 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.3 | 3.8 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 3.1 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 2.7 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.2 | 5.3 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 2.9 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 3.2 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 6.0 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.2 | 3.7 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 7.6 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 5.2 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 4.4 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 4.8 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.2 | 1.5 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 3.0 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.5 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 4.6 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 3.1 | REACTOME_ABC_FAMILY_PROTEINS_MEDIATED_TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 7.6 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.7 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 9.4 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 2.3 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.6 | REACTOME_E2F_ENABLED_INHIBITION_OF_PRE_REPLICATION_COMPLEX_FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 7.0 | REACTOME_ASPARAGINE_N_LINKED_GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.1 | 1.2 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 1.1 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 3.5 | REACTOME_CDK_MEDIATED_PHOSPHORYLATION_AND_REMOVAL_OF_CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 1.6 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.5 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.1 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.8 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 1.5 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.8 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.5 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.4 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.1 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.0 | REACTOME_CIRCADIAN_CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.6 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME_ER_PHAGOSOME_PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.2 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |