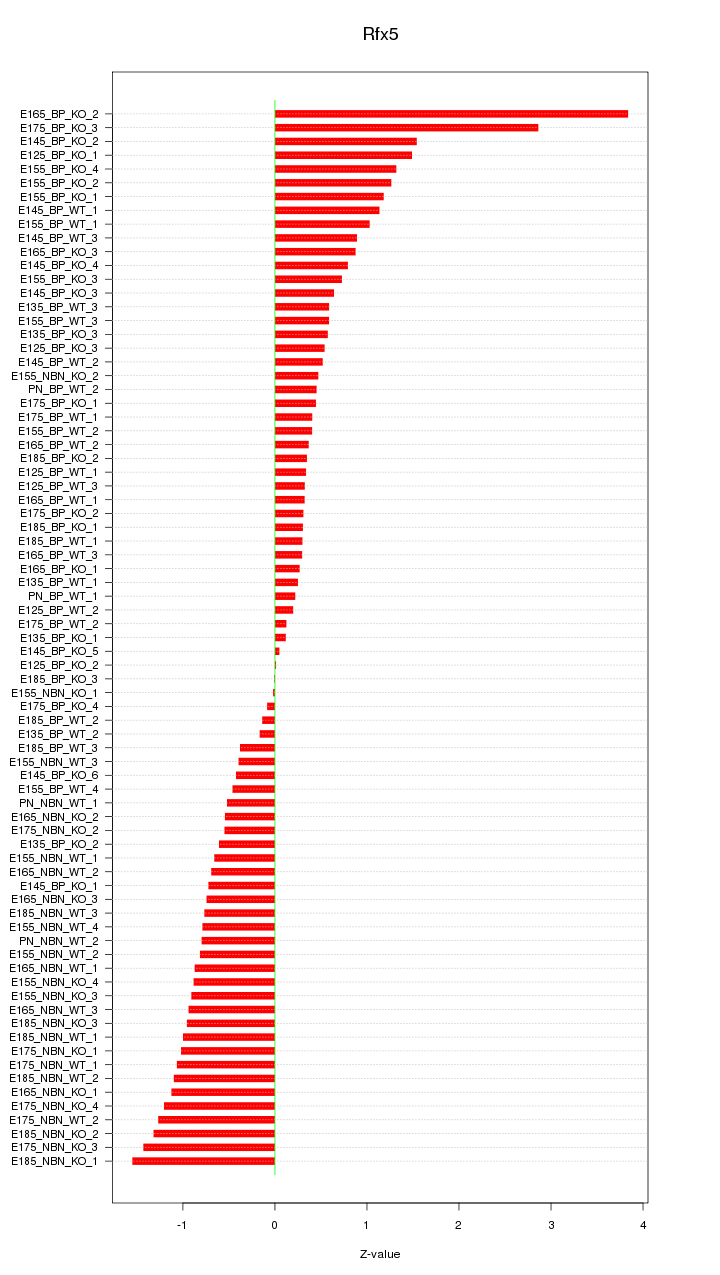
Motif ID: Rfx5
Z-value: 0.933

Transcription factors associated with Rfx5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rfx5 | ENSMUSG00000005774.6 | Rfx5 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx5 | mm10_v2_chr3_+_94954075_94954226 | 0.44 | 6.8e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.7 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 1.7 | 8.7 | GO:0002339 | B cell selection(GO:0002339) |
| 1.5 | 4.4 | GO:0070428 | granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 1.3 | 5.2 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 1.1 | 10.9 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.9 | 4.6 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.8 | 1.7 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.6 | 5.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.4 | 4.7 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.4 | 5.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.4 | 3.5 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.4 | 1.5 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.3 | 2.8 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.3 | 5.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.3 | 2.4 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.3 | 9.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.2 | 6.4 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.2 | 2.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.2 | 7.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 1.6 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.2 | 7.2 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 2.0 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.1 | 1.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 2.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 3.8 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 1.0 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 3.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.2 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.3 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 1.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.1 | GO:0007338 | single fertilization(GO:0007338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 22.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 1.8 | 7.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.4 | 5.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.1 | 13.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 1.0 | 1.0 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.4 | 3.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.4 | 1.6 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.3 | 5.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 1.7 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 3.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 10.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 2.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 5.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 1.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.2 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.1 | 4.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 3.2 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 2.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 15.0 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 10.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.6 | 6.4 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 1.3 | 3.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.8 | 8.7 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.6 | 9.6 | GO:0046977 | TAP binding(GO:0046977) |
| 0.6 | 4.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 2.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.4 | 5.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.3 | 7.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 8.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 3.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 2.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 2.3 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.2 | 4.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 7.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.4 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.1 | 2.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 1.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 14.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.7 | GO:0030145 | manganese ion binding(GO:0030145) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 4.2 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.0 | 3.5 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.5 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.6 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | REACTOME_ENDOSOMAL_VACUOLAR_PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 8.3 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.2 | 3.5 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 2.0 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 7.5 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 4.6 | REACTOME_REGULATION_OF_GLUCOKINASE_BY_GLUCOKINASE_REGULATORY_PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 4.4 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 6.4 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.7 | REACTOME_ER_PHAGOSOME_PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 10.7 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 5.5 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.4 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 1.6 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME_AUTODEGRADATION_OF_CDH1_BY_CDH1_APC_C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |