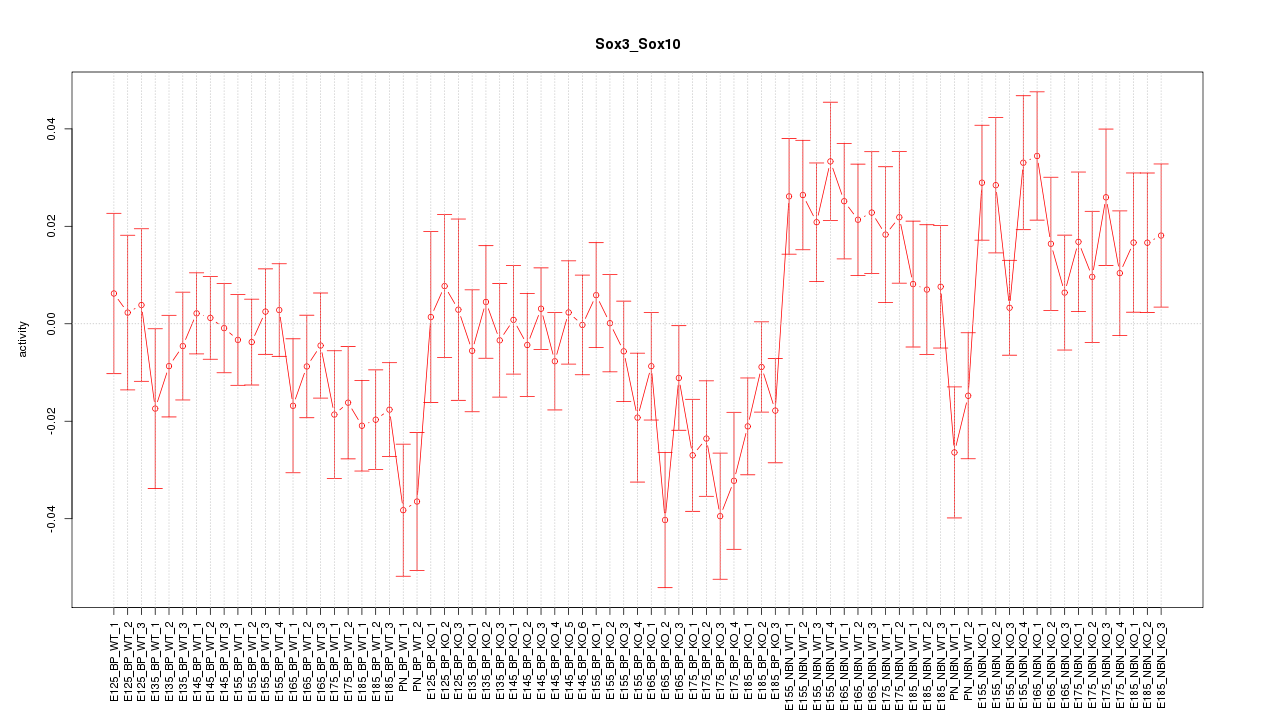
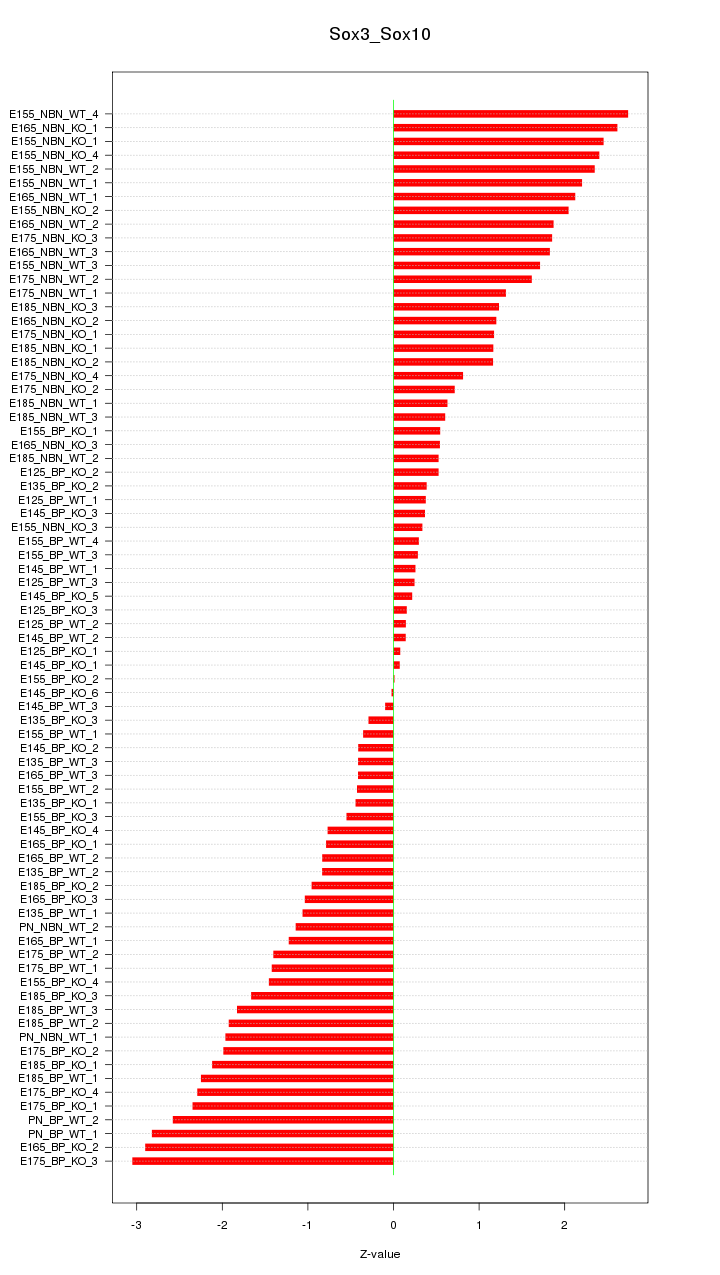
Motif ID: Sox3_Sox10
Z-value: 1.443

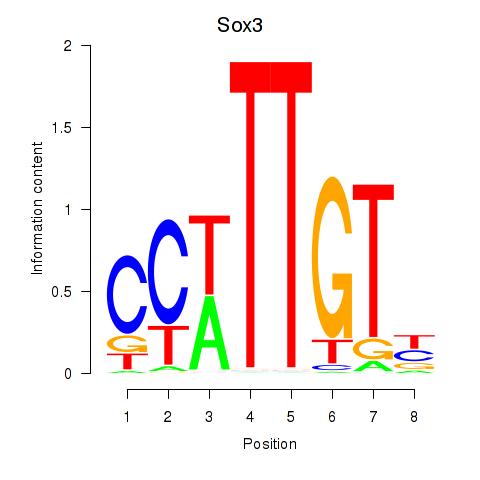
Transcription factors associated with Sox3_Sox10:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sox10 | ENSMUSG00000033006.9 | Sox10 |
| Sox3 | ENSMUSG00000045179.8 | Sox3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox3 | mm10_v2_chrX_-_60893430_60893440 | -0.17 | 1.5e-01 | Click! |
| Sox10 | mm10_v2_chr15_-_79164477_79164496 | 0.01 | 9.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 24.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 4.9 | 14.7 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 4.7 | 23.7 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 4.3 | 12.8 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 4.3 | 17.0 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 3.7 | 11.1 | GO:0007521 | muscle cell fate determination(GO:0007521) cellular response to parathyroid hormone stimulus(GO:0071374) |
| 3.6 | 14.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 3.5 | 24.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 3.3 | 10.0 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 3.3 | 29.7 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 3.2 | 9.7 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 2.7 | 35.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 2.6 | 2.6 | GO:0070428 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 2.6 | 13.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 2.3 | 22.9 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 2.2 | 10.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 2.0 | 21.9 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 1.9 | 15.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 1.9 | 7.6 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 1.9 | 9.3 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 1.9 | 16.7 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 1.7 | 5.1 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 1.7 | 5.1 | GO:1904207 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 1.7 | 5.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 1.6 | 24.2 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 1.6 | 11.1 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 1.6 | 6.3 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 1.5 | 12.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 1.5 | 16.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 1.5 | 5.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.4 | 2.8 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 1.4 | 5.6 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 1.4 | 4.2 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 1.4 | 9.8 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.4 | 4.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 1.3 | 4.0 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 1.3 | 5.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 1.3 | 7.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 1.3 | 10.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 1.3 | 7.6 | GO:0086018 | SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) |
| 1.3 | 3.8 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 1.2 | 3.7 | GO:0010814 | neuropeptide catabolic process(GO:0010813) substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.2 | 3.6 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 1.2 | 3.5 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 1.2 | 5.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 1.1 | 4.5 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 1.1 | 15.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 1.1 | 2.3 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 1.1 | 20.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 1.1 | 7.4 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 1.0 | 3.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 1.0 | 3.1 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 1.0 | 4.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 1.0 | 4.0 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.0 | 6.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 1.0 | 4.9 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.0 | 14.7 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 1.0 | 2.9 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 1.0 | 4.9 | GO:0017085 | response to insecticide(GO:0017085) |
| 1.0 | 8.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.0 | 3.8 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.0 | 1.0 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 0.9 | 2.8 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 0.9 | 4.6 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.9 | 3.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.9 | 6.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.9 | 7.2 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.9 | 2.7 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.9 | 9.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.9 | 2.7 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.9 | 5.2 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.8 | 4.2 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.8 | 2.5 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.8 | 2.5 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.8 | 2.5 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.8 | 7.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.8 | 19.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.8 | 7.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.8 | 4.0 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.8 | 4.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.7 | 2.2 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.7 | 4.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.7 | 6.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.7 | 2.8 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.7 | 8.5 | GO:0045760 | positive regulation of action potential(GO:0045760) |
| 0.7 | 4.9 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.7 | 2.1 | GO:0061744 | motor behavior(GO:0061744) |
| 0.7 | 5.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.7 | 4.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.7 | 4.0 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.7 | 4.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.7 | 2.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.7 | 5.9 | GO:0014848 | urinary bladder smooth muscle contraction(GO:0014832) urinary tract smooth muscle contraction(GO:0014848) |
| 0.6 | 1.9 | GO:0050883 | regulation of sulfur amino acid metabolic process(GO:0031335) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.6 | 3.1 | GO:0001661 | conditioned taste aversion(GO:0001661) amygdala development(GO:0021764) |
| 0.6 | 2.5 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.6 | 0.6 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.6 | 25.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.6 | 1.8 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.6 | 5.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.6 | 6.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.6 | 1.8 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.6 | 1.8 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.6 | 3.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.6 | 1.7 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.6 | 2.9 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.6 | 20.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.6 | 2.2 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.5 | 2.7 | GO:0031133 | cellular magnesium ion homeostasis(GO:0010961) regulation of axon diameter(GO:0031133) |
| 0.5 | 6.0 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.5 | 4.4 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.5 | 3.8 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.5 | 1.6 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.5 | 2.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.5 | 2.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.5 | 0.5 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.5 | 1.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.5 | 1.5 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.5 | 7.1 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.5 | 5.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.5 | 1.5 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.5 | 3.5 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.5 | 1.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.5 | 1.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.5 | 3.3 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.5 | 0.9 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.5 | 1.8 | GO:0016128 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.5 | 3.7 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.5 | 3.7 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.5 | 11.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.5 | 6.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.5 | 1.4 | GO:1900365 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.4 | 2.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 2.6 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.4 | 3.8 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.4 | 1.3 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.4 | 1.7 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.4 | 1.6 | GO:0051036 | regulation of endosome size(GO:0051036) |
| 0.4 | 1.2 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.4 | 13.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.4 | 1.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.4 | 1.2 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.4 | 9.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.4 | 0.8 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.4 | 7.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 1.2 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.4 | 1.2 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.4 | 5.3 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.4 | 1.5 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) negative regulation of PERK-mediated unfolded protein response(GO:1903898) positive regulation of receptor catabolic process(GO:2000646) |
| 0.4 | 1.1 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.4 | 1.8 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.4 | 1.8 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.4 | 3.3 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.4 | 3.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.4 | 1.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.3 | 4.5 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.3 | 0.7 | GO:0009838 | abscission(GO:0009838) |
| 0.3 | 5.1 | GO:0031111 | negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.3 | 54.9 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.3 | 5.0 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) |
| 0.3 | 1.7 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.3 | 2.6 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 | 1.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 1.6 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.3 | 24.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 2.9 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.3 | 10.5 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.3 | 0.3 | GO:0034767 | positive regulation of ion transmembrane transport(GO:0034767) |
| 0.3 | 1.6 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.3 | 1.3 | GO:2000858 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) positive regulation of mineralocorticoid secretion(GO:2000857) regulation of aldosterone secretion(GO:2000858) positive regulation of aldosterone secretion(GO:2000860) |
| 0.3 | 0.9 | GO:0035547 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.3 | 1.5 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.3 | 2.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.3 | 0.9 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.3 | 0.6 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.3 | 0.9 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 | 1.7 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.3 | 5.1 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.3 | 2.8 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.3 | 2.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.3 | 0.8 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 | 2.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.3 | 10.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.3 | 2.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.3 | 3.5 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.3 | 0.5 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.3 | 0.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.3 | 6.9 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.3 | 1.3 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.3 | 1.8 | GO:0021684 | cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 2.0 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.2 | 0.7 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.2 | 2.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 1.7 | GO:1903748 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) |
| 0.2 | 1.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.2 | 1.7 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 1.5 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.2 | 1.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 2.4 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.2 | 1.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.2 | 3.6 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.2 | 3.1 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.2 | 0.7 | GO:0031349 | positive regulation of defense response(GO:0031349) |
| 0.2 | 1.6 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.2 | 2.8 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.2 | 1.1 | GO:0045629 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 1.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.2 | 0.7 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.2 | 2.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.2 | 0.7 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 0.6 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.2 | 1.3 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.2 | 2.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 3.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 28.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.2 | 0.6 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 | 1.4 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.2 | 0.8 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.2 | 3.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.2 | 3.0 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 1.3 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 0.4 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.9 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 | 0.4 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.2 | 2.0 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.2 | 1.2 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.2 | 0.5 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 0.7 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.2 | 1.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.2 | 3.0 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 0.8 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.2 | 1.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.2 | 3.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.2 | 1.0 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.2 | 1.1 | GO:0003014 | renal system process(GO:0003014) |
| 0.2 | 1.4 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.2 | 0.8 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 | 0.5 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.2 | 0.9 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 1.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 6.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.3 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 1.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.9 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 1.1 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 2.0 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.6 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.7 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.1 | 3.8 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.1 | 0.7 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 | 1.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.7 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 3.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 5.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.9 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.1 | 3.4 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.1 | 0.5 | GO:0002339 | B cell selection(GO:0002339) |
| 0.1 | 0.5 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.1 | 0.5 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.4 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 0.4 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 1.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 2.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 1.1 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.1 | 0.9 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 1.5 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 1.4 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 1.0 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.7 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 3.3 | GO:0034763 | negative regulation of transmembrane transport(GO:0034763) |
| 0.1 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 4.0 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.1 | 4.1 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 0.7 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 0.2 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.1 | 0.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 1.1 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 11.6 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 1.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.3 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.1 | 1.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.4 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 6.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 1.7 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 1.0 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.7 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.1 | 2.7 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.9 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 1.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 1.0 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 | 1.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 1.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.3 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 2.1 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.2 | GO:1902950 | regulation of dendritic spine maintenance(GO:1902950) |
| 0.1 | 0.9 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.3 | GO:0035826 | rubidium ion transport(GO:0035826) cellular hypotonic response(GO:0071476) |
| 0.1 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.4 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 3.8 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.1 | 2.0 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.1 | 0.7 | GO:0014742 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.1 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 12.6 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.1 | 0.3 | GO:0001302 | replicative cell aging(GO:0001302) |
| 0.1 | 0.7 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.4 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.4 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.2 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 0.3 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.2 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.1 | 0.7 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 4.4 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.1 | 0.8 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 2.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.3 | GO:0010878 | cholesterol storage(GO:0010878) steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.1 | GO:1901526 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 6.5 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 2.0 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.3 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 4.3 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 11.2 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.8 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.6 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.6 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.5 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 1.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.8 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.7 | GO:0042439 | ethanolamine-containing compound metabolic process(GO:0042439) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.6 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 2.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.7 | GO:0061136 | regulation of proteasomal protein catabolic process(GO:0061136) |
| 0.0 | 0.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 6.6 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process(GO:0043161) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.7 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 5.7 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 1.0 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.4 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.0 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.4 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.5 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.7 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.2 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 3.8 | 38.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 2.7 | 24.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 2.2 | 4.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 2.1 | 51.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 2.0 | 4.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 1.8 | 1.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 1.7 | 26.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 1.4 | 4.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.4 | 56.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 1.3 | 5.3 | GO:0090537 | CERF complex(GO:0090537) |
| 1.2 | 5.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.1 | 17.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.1 | 6.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 1.1 | 5.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 1.0 | 18.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 1.0 | 7.7 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.9 | 7.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.9 | 41.3 | GO:0043034 | costamere(GO:0043034) |
| 0.8 | 12.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.8 | 4.0 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.7 | 11.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.7 | 3.0 | GO:0031673 | H zone(GO:0031673) |
| 0.6 | 40.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.6 | 7.3 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.5 | 1.6 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.5 | 8.7 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.5 | 2.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.5 | 2.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.5 | 1.5 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.5 | 2.0 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.5 | 1.0 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.5 | 6.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.5 | 70.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.5 | 4.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.4 | 3.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.4 | 1.3 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.4 | 1.3 | GO:0031533 | mRNA cap methyltransferase complex(GO:0031533) |
| 0.4 | 6.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.4 | 4.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 3.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 0.8 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.4 | 1.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.4 | 16.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 33.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.4 | 2.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.4 | 6.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.3 | 1.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 3.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 1.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.3 | 1.9 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.3 | 1.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.3 | 1.5 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.3 | 2.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.3 | 1.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 3.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 2.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 1.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 3.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.3 | 2.4 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.3 | 3.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 2.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 2.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 1.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 1.0 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.2 | 25.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 5.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 0.7 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 4.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 1.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 3.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 2.6 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 10.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 8.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 1.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 2.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 1.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 2.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 4.8 | GO:0044298 | cell body membrane(GO:0044298) |
| 0.2 | 9.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 2.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 3.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 1.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 0.7 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 2.6 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.2 | 0.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 4.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 0.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.3 | GO:0071014 | U2-type post-mRNA release spliceosomal complex(GO:0071008) post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 10.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 10.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.7 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 8.3 | GO:0015630 | microtubule cytoskeleton(GO:0015630) |
| 0.1 | 0.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.6 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 1.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.8 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.7 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.1 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 1.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.1 | 0.6 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 1.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 3.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 3.0 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 4.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 6.3 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 1.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 3.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 2.2 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 0.5 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 2.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 60.1 | GO:0005768 | endosome(GO:0005768) |
| 0.1 | 1.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 2.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 17.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 0.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.5 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 2.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.8 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 4.6 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 0.8 | GO:0099512 | supramolecular fiber(GO:0099512) polymeric cytoskeletal fiber(GO:0099513) |
| 0.1 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 4.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 5.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 2.4 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 12.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.7 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 1.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 29.7 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 3.8 | 18.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 2.9 | 23.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 2.2 | 24.4 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 2.2 | 24.4 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 2.2 | 10.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 2.1 | 6.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 2.1 | 8.6 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 2.1 | 17.0 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 1.9 | 5.7 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 1.8 | 10.8 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 1.8 | 5.3 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 1.7 | 23.8 | GO:0031005 | filamin binding(GO:0031005) |
| 1.7 | 5.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 1.5 | 9.3 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 1.5 | 6.0 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.5 | 5.8 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.4 | 4.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 1.3 | 11.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 1.2 | 3.6 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 1.2 | 38.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 1.2 | 4.6 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 1.1 | 7.6 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.1 | 5.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.1 | 5.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.0 | 3.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.0 | 3.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 1.0 | 4.0 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.9 | 13.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.9 | 59.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.9 | 1.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.9 | 2.7 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.9 | 6.8 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.8 | 2.5 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.8 | 4.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.8 | 4.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.8 | 7.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.8 | 7.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.8 | 3.8 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.7 | 2.9 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.7 | 3.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.7 | 19.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.7 | 2.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.7 | 5.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.7 | 2.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.6 | 1.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.6 | 21.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.6 | 5.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 5.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.6 | 2.5 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.6 | 2.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.6 | 1.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.6 | 9.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.6 | 1.7 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.6 | 7.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.6 | 8.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.5 | 6.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.5 | 3.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.5 | 6.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.5 | 5.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.5 | 6.7 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.5 | 1.8 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.5 | 9.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.4 | 9.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.4 | 16.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.4 | 1.3 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.4 | 1.6 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 4.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.4 | 3.6 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.4 | 1.6 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.4 | 22.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.4 | 1.2 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.4 | 2.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.4 | 5.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.4 | 2.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.4 | 2.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.4 | 3.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.4 | 7.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.4 | 1.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.4 | 0.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.4 | 4.4 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.4 | 2.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 2.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.3 | 3.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 6.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.3 | 5.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 13.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.3 | 1.0 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.3 | 3.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.3 | 10.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 1.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.3 | 4.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 0.9 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.3 | 1.8 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.3 | 2.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.3 | 1.5 | GO:0043559 | insulin binding(GO:0043559) |
| 0.3 | 0.9 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.3 | 23.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 3.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 15.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.3 | 0.8 | GO:0004534 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) 5'-3' exoribonuclease activity(GO:0004534) |
| 0.3 | 5.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 4.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.3 | 2.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 27.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 19.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.2 | 38.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 6.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 0.7 | GO:0015173 | hydrogen:amino acid symporter activity(GO:0005280) aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.2 | 2.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 4.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 7.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.2 | 1.2 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 7.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 0.7 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.2 | 1.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 1.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 3.0 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 7.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 1.0 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.2 | 4.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 2.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 51.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.2 | 4.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.2 | 7.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 1.6 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 1.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 2.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 11.0 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.2 | 0.6 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 2.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 1.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 3.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.2 | 2.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 20.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.2 | 0.9 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.2 | 1.6 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.2 | 1.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 38.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.2 | 3.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 2.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.2 | 1.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 1.7 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 2.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 2.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 4.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.1 | 0.7 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 2.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 2.8 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 2.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.7 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.1 | 1.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.5 | GO:0071532 | ornithine decarboxylase inhibitor activity(GO:0008073) ankyrin repeat binding(GO:0071532) |
| 0.1 | 3.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.7 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.1 | 1.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 2.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 6.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 2.2 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 4.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.7 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 2.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 5.5 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 1.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.6 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 13.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 3.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 11.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 9.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 1.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.7 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 17.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 1.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.4 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 5.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.8 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 2.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 2.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 0.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.5 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 1.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 2.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 2.0 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 6.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 1.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 3.6 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0098505 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.5 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.0 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 30.9 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.7 | 9.1 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.6 | 17.5 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.6 | 16.9 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.4 | 31.5 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.4 | 30.9 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.4 | 15.1 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.4 | 10.6 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.3 | 12.0 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 10.8 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.3 | 7.4 | PID_ER_NONGENOMIC_PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.3 | 13.0 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.3 | 12.7 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 13.8 | PID_INSULIN_PATHWAY | Insulin Pathway |
| 0.2 | 1.4 | PID_S1P_S1P2_PATHWAY | S1P2 pathway |
| 0.2 | 7.3 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 8.3 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.2 | 3.4 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 9.5 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.2 | 13.6 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.2 | 6.0 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 4.0 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.2 | 8.2 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.2 | 19.2 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 2.5 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 3.9 | ST_G_ALPHA_I_PATHWAY | G alpha i Pathway |
| 0.1 | 2.7 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 1.1 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 10.0 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.1 | 1.2 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.7 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.5 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 13.6 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 3.1 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.1 | 1.2 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.9 | PID_IFNG_PATHWAY | IFN-gamma pathway |
| 0.1 | 2.2 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.1 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.6 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.6 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.1 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 0.3 | ST_JAK_STAT_PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.2 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.5 | PID_ERBB1_DOWNSTREAM_PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.3 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.2 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID_PI3KCI_PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.6 | PID_NFAT_3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.5 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 25.8 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 1.2 | 27.9 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.2 | 6.0 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 1.1 | 16.8 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 1.0 | 43.7 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 1.0 | 21.5 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 1.0 | 10.0 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.8 | 10.8 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.8 | 12.7 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.7 | 2.8 | REACTOME_FGFR2C_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.7 | 15.0 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.6 | 16.8 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.5 | 7.3 | REACTOME_INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.5 | 12.4 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.5 | 3.7 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.5 | 8.1 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.4 | 11.0 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 15.9 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.4 | 5.7 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_LATE_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 4.8 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 5.7 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 11.0 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.3 | 4.6 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 25.5 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 5.8 | REACTOME_REGULATION_OF_KIT_SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.3 | 13.4 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 2.2 | REACTOME_ARMS_MEDIATED_ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.3 | 6.5 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 9.2 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 0.9 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.3 | 5.6 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.3 | 2.1 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 7.1 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.3 | 2.6 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 17.0 | REACTOME_L1CAM_INTERACTIONS | Genes involved in L1CAM interactions |
| 0.3 | 26.3 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 9.4 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 10.4 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.2 | 24.9 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.2 | 6.0 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.2 | 2.7 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 1.5 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 2.6 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 1.8 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 8.4 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 0.6 | REACTOME_NEP_NS2_INTERACTS_WITH_THE_CELLULAR_EXPORT_MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 4.1 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.2 | 16.9 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 2.2 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 2.4 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 3.3 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 1.5 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.3 | REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.4 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.2 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 3.8 | REACTOME_REGULATION_OF_GLUCOKINASE_BY_GLUCOKINASE_REGULATORY_PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 3.0 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.3 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.3 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.7 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.6 | REACTOME_PHOSPHORYLATION_OF_THE_APC_C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 2.1 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.9 | REACTOME_TRANSMISSION_ACROSS_CHEMICAL_SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.1 | 1.3 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 2.5 | REACTOME_PROTEIN_FOLDING | Genes involved in Protein folding |
| 0.1 | 3.5 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 4.6 | REACTOME_TRANS_GOLGI_NETWORK_VESICLE_BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.1 | 0.4 | REACTOME_PD1_SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.9 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.6 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.2 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.7 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.1 | 2.9 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 0.5 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 1.2 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 3.1 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 3.0 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 4.7 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 3.0 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT_ATP_SYNTHESIS_BY_CHEMIOSMOTIC_COUPLING_AND_HEAT_PRODUCTION_BY_UNCOUPLING_PROTEINS_ | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 1.1 | REACTOME_MRNA_CAPPING | Genes involved in mRNA Capping |
| 0.0 | 2.7 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 3.8 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 3.2 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.0 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.0 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.5 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME_TRANSPORT_OF_MATURE_MRNA_DERIVED_FROM_AN_INTRONLESS_TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.1 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |