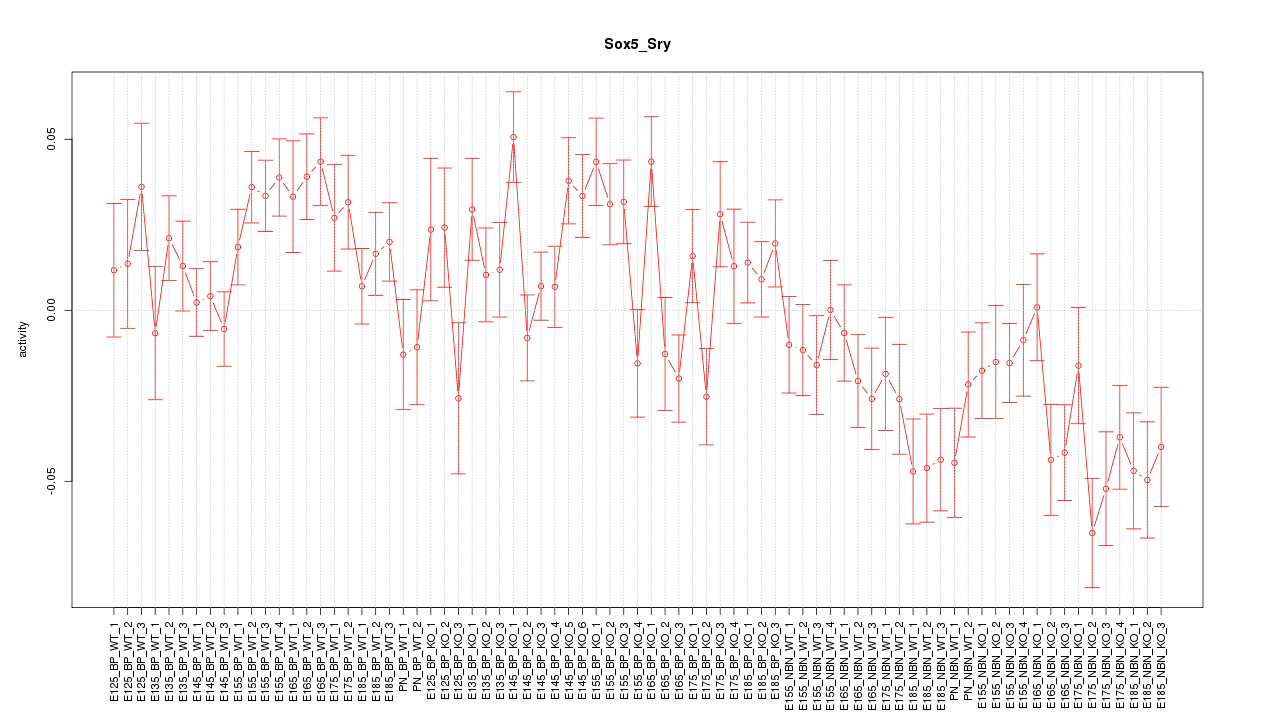
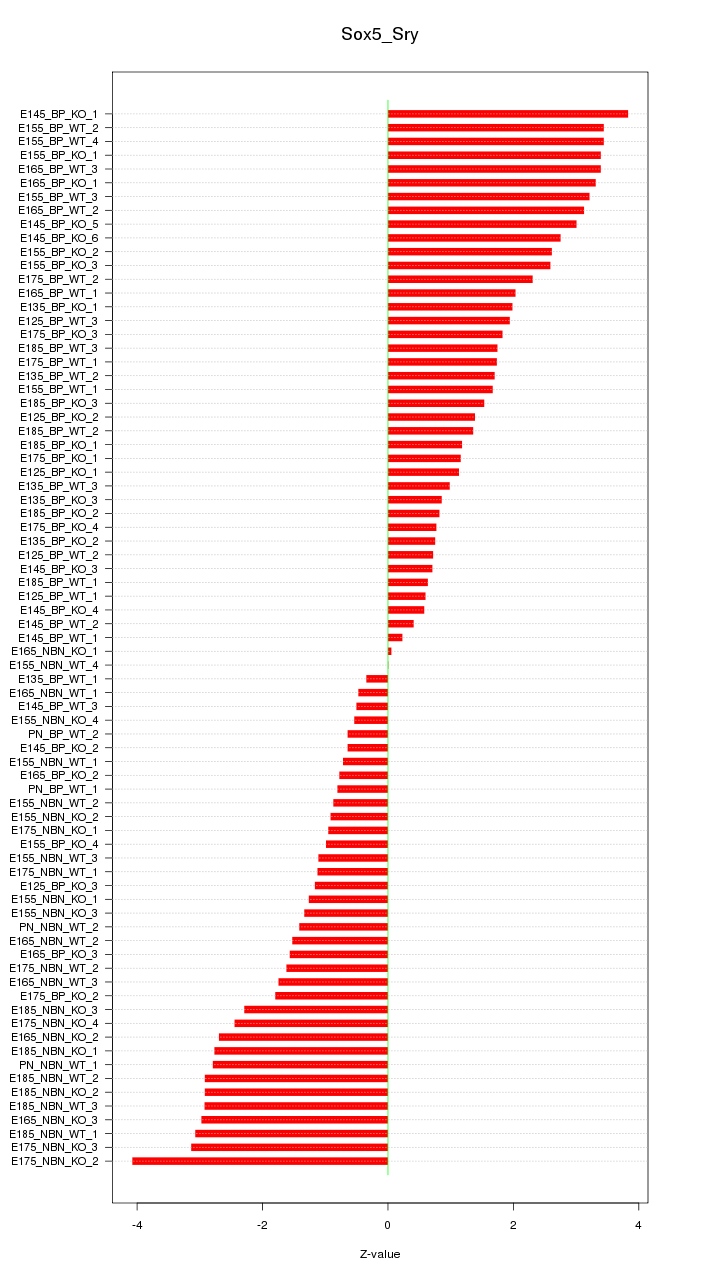
Motif ID: Sox5_Sry
Z-value: 1.991
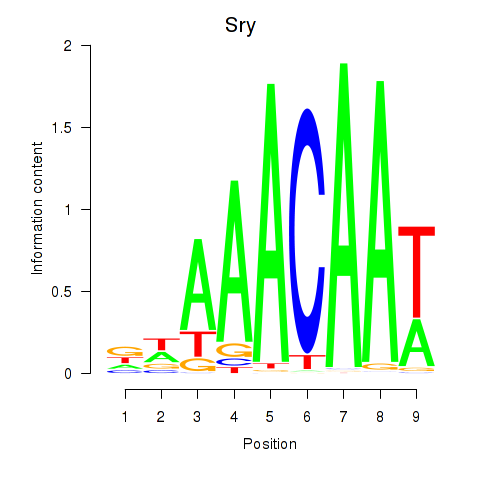
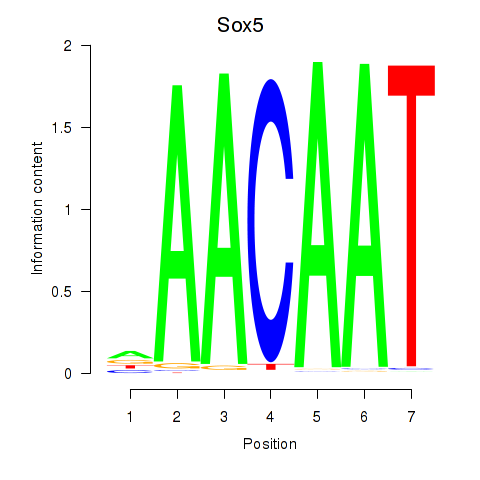
Transcription factors associated with Sox5_Sry:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sox5 | ENSMUSG00000041540.10 | Sox5 |
| Sry | ENSMUSG00000069036.3 | Sry |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox5 | mm10_v2_chr6_-_143947092_143947111 | 0.24 | 3.4e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.0 | 52.0 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 8.6 | 25.9 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 8.3 | 24.8 | GO:0060023 | soft palate development(GO:0060023) |
| 7.0 | 56.3 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 6.5 | 19.4 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 5.4 | 21.6 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 4.4 | 48.5 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 4.2 | 16.7 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 3.7 | 11.2 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 3.7 | 11.0 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 3.2 | 38.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 3.2 | 50.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 3.0 | 27.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 3.0 | 21.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 3.0 | 14.9 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 2.9 | 11.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 2.8 | 8.5 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 2.7 | 13.7 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 2.3 | 20.3 | GO:0030035 | microspike assembly(GO:0030035) |
| 2.1 | 15.0 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 2.0 | 11.8 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 1.9 | 9.7 | GO:0042891 | antibiotic transport(GO:0042891) dipeptide transport(GO:0042938) |
| 1.8 | 18.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 1.7 | 6.9 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 1.7 | 8.5 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 1.6 | 8.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 1.4 | 5.8 | GO:0061428 | embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 1.4 | 7.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 1.3 | 14.8 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 1.3 | 11.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 1.2 | 3.7 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 1.2 | 13.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 1.2 | 6.0 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 1.2 | 8.4 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 1.2 | 15.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 1.2 | 9.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 1.1 | 5.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 1.1 | 9.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 1.1 | 3.3 | GO:0003195 | tricuspid valve formation(GO:0003195) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 1.1 | 3.2 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 1.1 | 15.1 | GO:0072189 | ureter development(GO:0072189) |
| 1.1 | 16.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 1.0 | 14.6 | GO:0060013 | righting reflex(GO:0060013) |
| 1.0 | 9.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 1.0 | 3.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 1.0 | 7.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 1.0 | 4.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 1.0 | 11.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 1.0 | 2.9 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.9 | 8.4 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.9 | 7.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.9 | 1.7 | GO:0003266 | regulation of secondary heart field cardioblast proliferation(GO:0003266) |
| 0.9 | 6.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.9 | 3.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.8 | 2.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.8 | 7.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.8 | 2.4 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.8 | 5.6 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.8 | 2.4 | GO:0046544 | regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) development of secondary male sexual characteristics(GO:0046544) |
| 0.8 | 4.7 | GO:0048840 | otolith development(GO:0048840) |
| 0.8 | 5.4 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.8 | 1.5 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.7 | 8.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.7 | 0.7 | GO:0003263 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) |
| 0.7 | 4.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.6 | 6.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.6 | 4.9 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.6 | 4.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.6 | 2.8 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.5 | 1.6 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.5 | 9.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.5 | 4.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.5 | 1.0 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.5 | 2.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.5 | 3.4 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.5 | 6.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.5 | 1.4 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.5 | 1.9 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.5 | 4.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.4 | 1.8 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.4 | 1.7 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.4 | 12.9 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.4 | 2.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.4 | 10.0 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.4 | 2.0 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.4 | 4.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.4 | 1.9 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.4 | 1.9 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.4 | 1.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.4 | 2.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.3 | 1.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 1.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.3 | 1.3 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.3 | 29.8 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.3 | 0.6 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.3 | 2.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.3 | 3.8 | GO:0060004 | reflex(GO:0060004) |
| 0.3 | 1.2 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.3 | 1.2 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.3 | 15.4 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.3 | 2.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.3 | 3.0 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.3 | 4.8 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.3 | 1.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 1.5 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.3 | 3.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.2 | 0.5 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.2 | 3.9 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 4.9 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 1.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.2 | 1.6 | GO:0016071 | mRNA metabolic process(GO:0016071) |
| 0.2 | 1.2 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.2 | 1.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 0.5 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 5.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 4.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 1.3 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.2 | 9.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.2 | 0.4 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 3.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 6.1 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.2 | 5.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 13.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 2.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 4.3 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.2 | 5.8 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.2 | 0.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 2.7 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 5.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.1 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 0.4 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.1 | 1.1 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 1.9 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 9.9 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.1 | 12.8 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.1 | 0.6 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.1 | 0.8 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.1 | 10.1 | GO:0048469 | cell maturation(GO:0048469) |
| 0.1 | 3.8 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.1 | 1.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 2.0 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 1.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 2.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.8 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 8.0 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.1 | 1.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.8 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.5 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.5 | GO:0042759 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.7 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 2.1 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.0 | 0.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.9 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) small RNA loading onto RISC(GO:0070922) |
| 0.0 | 0.1 | GO:0060686 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 2.1 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.4 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.7 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.5 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.5 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 3.7 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.0 | 1.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 14.9 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 2.5 | 52.0 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 2.2 | 11.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 1.8 | 16.5 | GO:0030478 | actin cap(GO:0030478) |
| 1.5 | 23.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.4 | 7.0 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.2 | 3.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 1.2 | 9.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 1.0 | 4.2 | GO:0031673 | H zone(GO:0031673) |
| 0.9 | 7.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.8 | 9.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.8 | 2.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.7 | 3.6 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.7 | 13.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.7 | 21.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.5 | 6.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.5 | 31.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.5 | 3.4 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.5 | 9.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.4 | 3.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 17.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.4 | 1.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 1.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.3 | 54.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.3 | 49.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.3 | 3.8 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.3 | 10.5 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.3 | 4.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 4.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 26.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 20.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 2.5 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.2 | 1.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 1.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 3.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 14.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 1.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 9.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 27.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 10.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 6.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.5 | GO:0099569 | cone cell pedicle(GO:0044316) presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 1.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 4.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 39.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 17.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 3.6 | GO:0032153 | cell division site(GO:0032153) cell division site part(GO:0032155) |
| 0.1 | 1.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 49.3 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.1 | 3.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 7.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 6.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 4.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 2.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 2.1 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 3.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 5.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 161.1 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.4 | GO:0043197 | dendritic spine(GO:0043197) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 53.7 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 3.5 | 21.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 3.3 | 16.7 | GO:0043532 | angiostatin binding(GO:0043532) |
| 3.2 | 9.7 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 2.4 | 16.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 2.3 | 9.2 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 2.3 | 11.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 1.9 | 70.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 1.8 | 7.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 1.7 | 24.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 1.7 | 3.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 1.6 | 13.1 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 1.4 | 27.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.4 | 25.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 1.3 | 29.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 1.3 | 19.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.3 | 58.2 | GO:0070888 | E-box binding(GO:0070888) |
| 1.3 | 1.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 1.2 | 4.7 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 1.0 | 14.9 | GO:0035173 | histone kinase activity(GO:0035173) |
| 1.0 | 19.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.9 | 5.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.9 | 32.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.6 | 2.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.6 | 8.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.6 | 32.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.6 | 11.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.5 | 89.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.5 | 2.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.5 | 8.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.5 | 11.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.5 | 7.0 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.5 | 6.0 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.5 | 6.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.4 | 5.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.4 | 3.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 11.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.4 | 2.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.4 | 4.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.4 | 1.9 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.4 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 4.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.3 | 1.3 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 1.6 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.3 | 8.9 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.3 | 4.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 4.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.3 | 2.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 1.9 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.3 | 1.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.3 | 2.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.3 | 1.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.2 | 2.5 | GO:0035005 | lipid kinase activity(GO:0001727) 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 4.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.2 | 3.4 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.2 | 36.9 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.2 | 0.5 | GO:0002135 | CTP binding(GO:0002135) |
| 0.2 | 2.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 1.3 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.2 | 1.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 5.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 20.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 3.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 6.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.2 | 3.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 1.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 3.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 2.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 4.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 2.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 7.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 2.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 3.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 6.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 0.6 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 1.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 2.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 32.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 19.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 6.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.7 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.4 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 2.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 2.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 1.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.7 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.1 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.8 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 1.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.0 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 16.7 | GO:0003677 | DNA binding(GO:0003677) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 80.3 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 1.1 | 14.9 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.9 | 55.1 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.7 | 63.1 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.4 | 23.1 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.4 | 9.8 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.3 | 13.7 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.3 | 34.1 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.3 | 8.5 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.3 | 15.1 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.3 | 7.8 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.3 | 3.1 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.3 | 18.2 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.3 | 14.3 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.3 | 33.1 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 1.0 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.2 | 5.8 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 15.4 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.2 | 1.3 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.2 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 10.1 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.2 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 6.0 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.1 | 7.3 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.4 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 3.7 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.6 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 14.8 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.5 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.3 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.7 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.3 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.1 | 0.8 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 52.0 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 1.2 | 9.2 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 1.2 | 19.6 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 1.1 | 40.4 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 1.0 | 10.5 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 7.8 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.8 | 27.8 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.6 | 16.0 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.6 | 17.8 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.6 | 31.4 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.5 | 12.0 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.4 | 14.8 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.4 | 13.3 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 4.6 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.3 | 19.0 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 2.4 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 23.0 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 30.5 | REACTOME_MEIOSIS | Genes involved in Meiosis |
| 0.2 | 17.2 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 12.8 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 3.0 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 1.4 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.2 | 2.1 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 3.2 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 11.0 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 5.4 | REACTOME_RECYCLING_PATHWAY_OF_L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.9 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 4.4 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.5 | REACTOME_L1CAM_INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 3.2 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.2 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.5 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.3 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.1 | 1.3 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.9 | REACTOME_NOD1_2_SIGNALING_PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 15.8 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.1 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.1 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.6 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 2.5 | REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 2.0 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.5 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.3 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.6 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.7 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME_RAF_MAP_KINASE_CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 2.0 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |