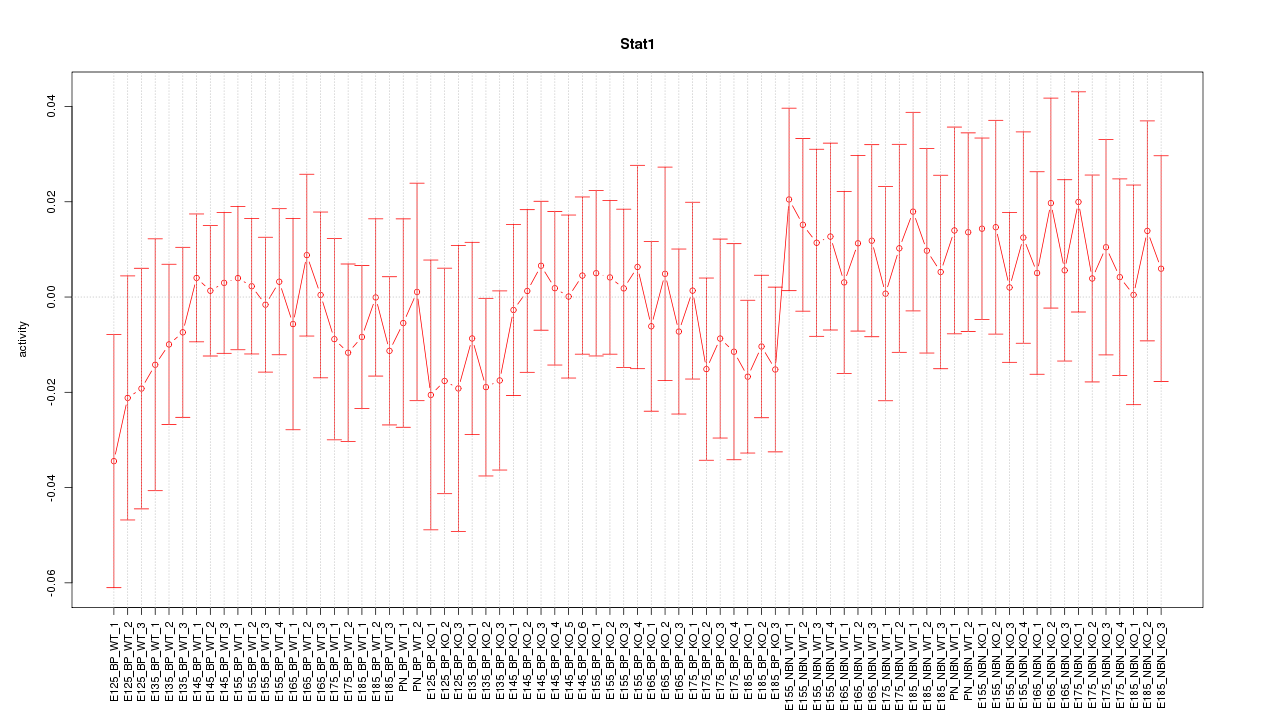
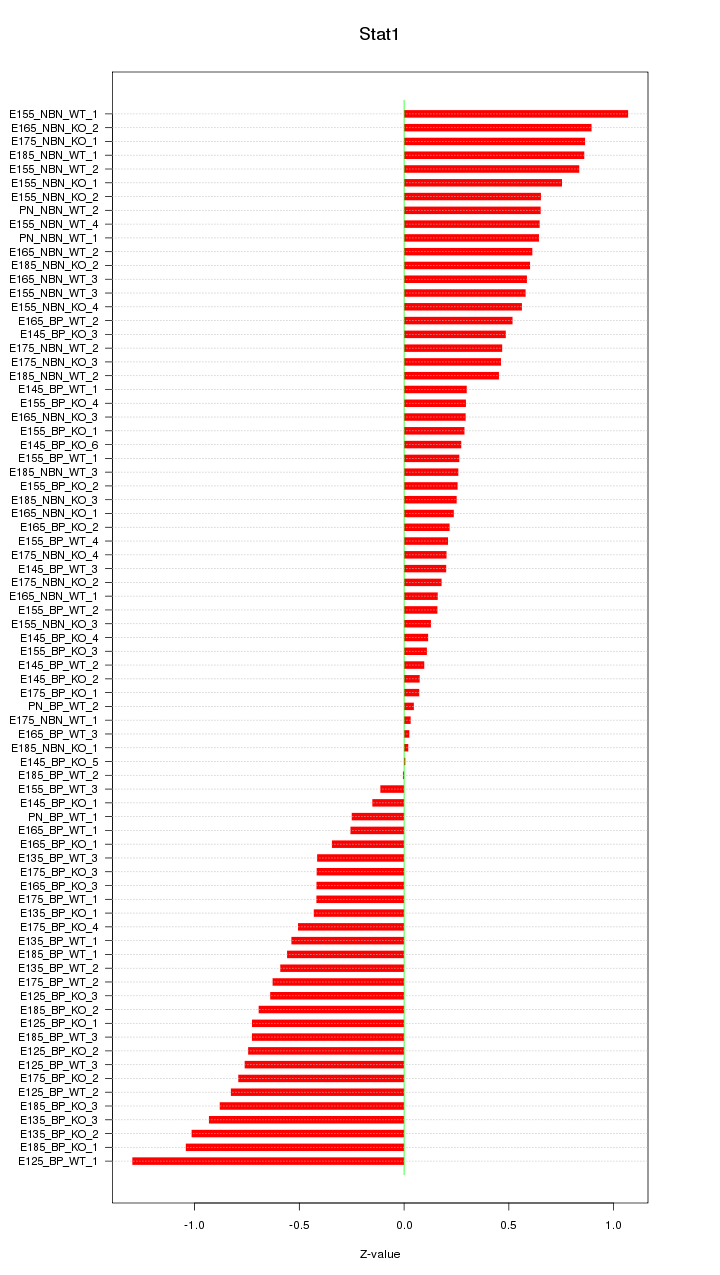
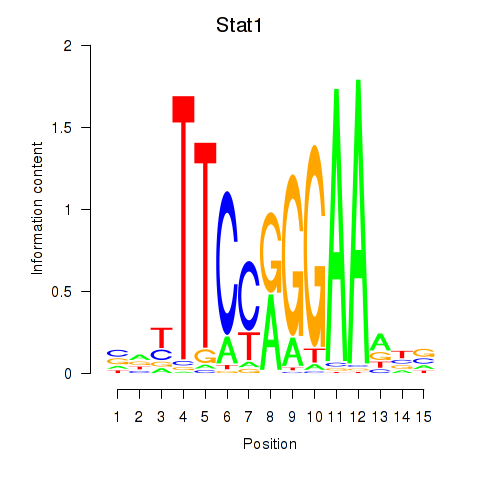
Motif ID: Stat1
Z-value: 0.546
Transcription factors associated with Stat1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Stat1 | ENSMUSG00000026104.8 | Stat1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat1 | mm10_v2_chr1_+_52119438_52119499 | -0.51 | 1.7e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 8.9 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 1.1 | 4.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 1.0 | 3.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 1.0 | 4.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.8 | 3.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.8 | 2.3 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) negative regulation of lymphocyte migration(GO:2000402) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.7 | 7.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) liver regeneration(GO:0097421) |
| 0.6 | 1.9 | GO:0002314 | germinal center B cell differentiation(GO:0002314) positive regulation of germinal center formation(GO:0002636) adenosine catabolic process(GO:0006154) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) purine nucleobase salvage(GO:0043096) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) xanthine metabolic process(GO:0046110) purine deoxyribonucleoside metabolic process(GO:0046122) regulation of adenosine receptor signaling pathway(GO:0060167) negative regulation of mucus secretion(GO:0070256) |
| 0.6 | 3.7 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.6 | 2.9 | GO:2000427 | eosinophil chemotaxis(GO:0048245) T cell extravasation(GO:0072683) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.5 | 3.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) phosphate ion transmembrane transport(GO:0035435) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.5 | 1.5 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.5 | 3.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.4 | 3.7 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.4 | 1.5 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.4 | 1.1 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.4 | 0.4 | GO:1903598 | positive regulation of gap junction assembly(GO:1903598) |
| 0.3 | 1.7 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.3 | 1.0 | GO:0001803 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.3 | 2.3 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.2 | 2.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 7.3 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.2 | 0.9 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 0.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 2.4 | GO:1901898 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 0.5 | GO:1904154 | trimming of terminal mannose on B branch(GO:0036509) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 1.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.2 | 0.8 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 2.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 3.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.6 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 3.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 1.0 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 2.4 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.1 | 8.5 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 0.8 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.3 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.5 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.5 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 | 1.7 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 2.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 5.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.6 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.6 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.2 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.1 | 1.6 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 1.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.3 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 1.6 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.4 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 1.7 | GO:0042993 | positive regulation of transcription factor import into nucleus(GO:0042993) |
| 0.0 | 0.7 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.7 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.8 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 2.3 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 1.5 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 1.0 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.6 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.0 | GO:2000564 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.6 | GO:0051591 | response to cAMP(GO:0051591) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.0 | 2.9 | GO:0044299 | C-fiber(GO:0044299) |
| 0.7 | 3.7 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.6 | 4.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.4 | 1.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.4 | 3.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 1.2 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 1.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.3 | 1.0 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 1.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 0.9 | GO:0090537 | CERF complex(GO:0090537) |
| 0.2 | 0.8 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 3.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.8 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 1.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.6 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 2.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 1.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 6.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.1 | 2.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 7.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.7 | GO:0097708 | cytoplasmic vesicle(GO:0031410) intracellular vesicle(GO:0097708) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 15.1 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.5 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 1.1 | 3.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 1.0 | 4.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.9 | 3.5 | GO:0019962 | interferon receptor activity(GO:0004904) type I interferon receptor activity(GO:0004905) type I interferon binding(GO:0019962) |
| 0.7 | 3.7 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.6 | 3.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.6 | 3.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.5 | 2.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.5 | 3.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 1.5 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.5 | 8.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.4 | 2.8 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.3 | 1.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.3 | 0.9 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.3 | 1.6 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.2 | 1.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 8.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.0 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.2 | 0.8 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 1.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 3.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 1.8 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 2.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.8 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.9 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 4.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.5 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 2.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 4.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.0 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 10.1 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.6 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.4 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 6.6 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.1 | 1.5 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.1 | 2.9 | PID_IL23_PATHWAY | IL23-mediated signaling events |
| 0.1 | 3.1 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.4 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.2 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.7 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.9 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.1 | 2.8 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 2.8 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.0 | 1.2 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.6 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.7 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.5 | 8.5 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 3.5 | REACTOME_REGULATION_OF_IFNA_SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 4.1 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 3.8 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 1.7 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 3.3 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 11.2 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.9 | REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 1.9 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.6 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.5 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.3 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.4 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.5 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.6 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 3.7 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.6 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 2.9 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.1 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 4.6 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME_RNA_POL_I_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.7 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.9 | REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.5 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |