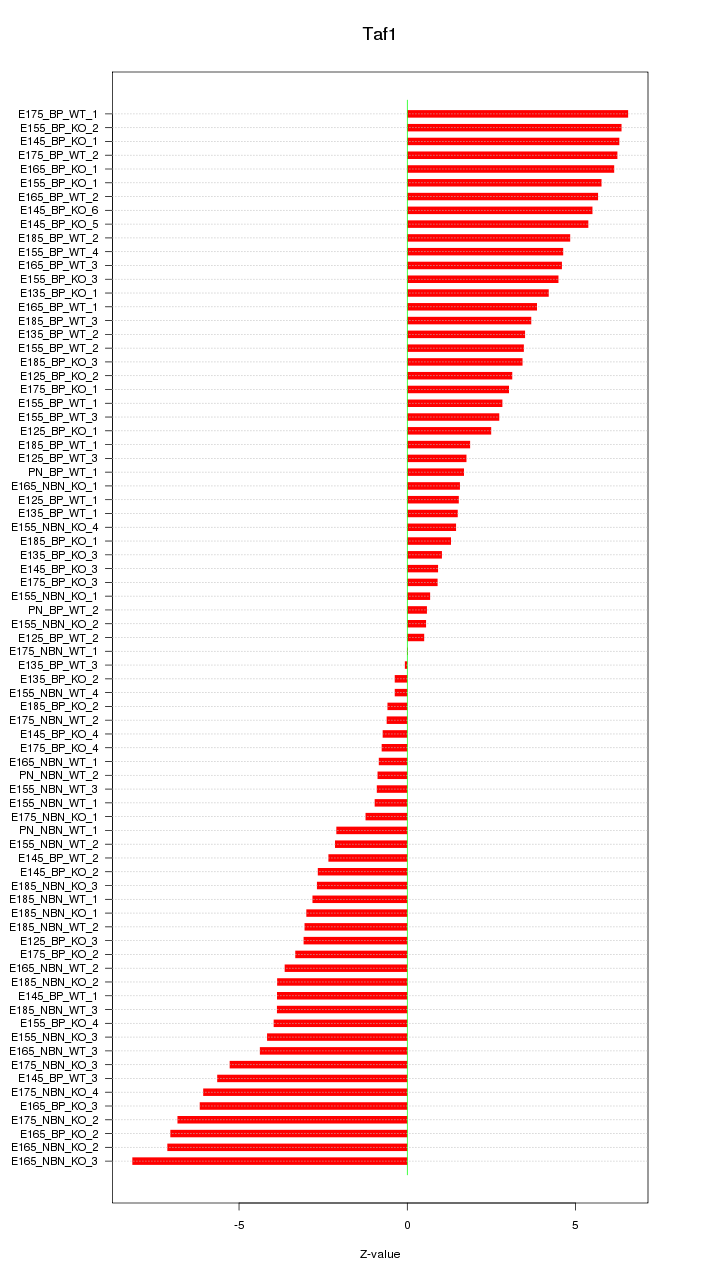
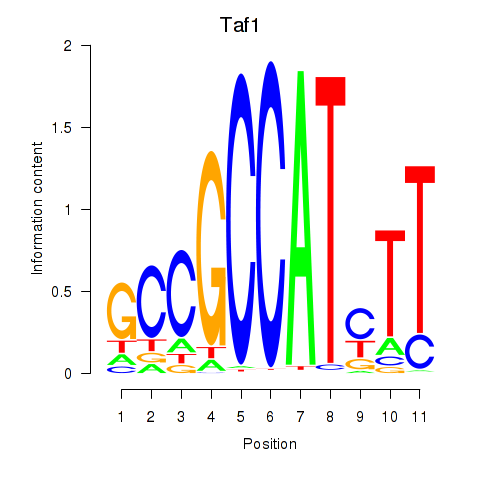
Motif ID: Taf1
Z-value: 3.780
Transcription factors associated with Taf1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Taf1 | ENSMUSG00000031314.11 | Taf1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Taf1 | mm10_v2_chrX_+_101532734_101532777 | 0.76 | 5.9e-16 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 44.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 7.6 | 61.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 7.5 | 29.9 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 7.1 | 28.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 6.8 | 20.4 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 6.8 | 20.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 6.7 | 33.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 6.3 | 19.0 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 6.3 | 31.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 6.1 | 18.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 5.8 | 17.4 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 5.8 | 28.9 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 5.6 | 16.7 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 5.3 | 53.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 5.3 | 21.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905024) |
| 5.0 | 20.0 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 4.8 | 33.9 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 4.7 | 18.9 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 4.7 | 14.0 | GO:0072554 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 4.6 | 13.9 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 4.5 | 45.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 4.5 | 13.6 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 4.4 | 22.2 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 4.1 | 8.2 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 3.9 | 11.8 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 3.9 | 15.6 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 3.8 | 15.0 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 3.7 | 22.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) establishment of Sertoli cell barrier(GO:0097368) |
| 3.6 | 14.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 3.6 | 10.7 | GO:0032240 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 3.5 | 10.6 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 3.5 | 17.6 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 3.5 | 17.6 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 3.5 | 3.5 | GO:0060699 | regulation of endoribonuclease activity(GO:0060699) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 3.4 | 13.5 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 3.3 | 6.7 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 3.3 | 33.0 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 3.2 | 22.3 | GO:0030242 | pexophagy(GO:0030242) |
| 3.2 | 12.6 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 3.1 | 12.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 3.1 | 9.3 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 3.1 | 34.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 3.1 | 9.3 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 3.0 | 12.0 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 2.9 | 11.7 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) |
| 2.9 | 20.4 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 2.9 | 8.6 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 2.8 | 8.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 2.8 | 22.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 2.7 | 8.2 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 2.7 | 8.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 2.7 | 24.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 2.7 | 8.1 | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 2.7 | 18.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 2.6 | 7.7 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 2.6 | 10.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 2.6 | 7.7 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 2.6 | 35.9 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 2.5 | 7.6 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 2.5 | 9.9 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 2.5 | 24.8 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 2.4 | 12.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 2.4 | 7.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 2.3 | 4.6 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 2.3 | 6.8 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 2.2 | 11.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 2.2 | 6.6 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 2.2 | 6.6 | GO:0019046 | release from viral latency(GO:0019046) |
| 2.2 | 12.9 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 2.1 | 8.6 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 2.1 | 8.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 2.1 | 6.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 2.0 | 12.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 2.0 | 10.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 2.0 | 15.9 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 2.0 | 5.9 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 1.9 | 7.8 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 1.9 | 5.8 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 1.9 | 15.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 1.9 | 13.4 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 1.9 | 13.4 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 1.9 | 26.5 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 1.8 | 11.0 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 1.8 | 12.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 1.8 | 5.5 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 1.8 | 9.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 1.8 | 5.4 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 1.8 | 9.0 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 1.8 | 19.6 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 1.8 | 14.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 1.7 | 10.4 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 1.7 | 5.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 1.7 | 1.7 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 1.7 | 5.2 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.7 | 5.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 1.7 | 10.2 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 1.7 | 6.8 | GO:0045358 | germ-line stem cell population maintenance(GO:0030718) negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 1.7 | 5.0 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 1.7 | 5.0 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 1.6 | 16.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 1.6 | 8.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 1.6 | 9.8 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 1.6 | 6.5 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 1.6 | 6.5 | GO:0097694 | establishment of RNA localization to telomere(GO:0097694) |
| 1.6 | 3.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 1.6 | 7.9 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 1.6 | 15.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 1.5 | 4.6 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 1.5 | 7.7 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 1.5 | 6.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 1.5 | 6.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 1.5 | 5.9 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 1.5 | 10.3 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 1.5 | 8.8 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 1.5 | 5.9 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 1.4 | 4.3 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 1.4 | 13.0 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 1.4 | 10.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 1.4 | 4.2 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 1.4 | 4.2 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 1.4 | 9.8 | GO:0090169 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 1.4 | 14.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 1.4 | 14.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 1.4 | 4.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 1.4 | 16.6 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 1.3 | 9.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 1.3 | 9.3 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 1.3 | 6.5 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 1.3 | 7.8 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 1.3 | 5.0 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 1.3 | 6.3 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 1.3 | 7.5 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 1.2 | 6.2 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.2 | 22.3 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 1.2 | 7.4 | GO:0021631 | optic nerve morphogenesis(GO:0021631) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 1.2 | 4.9 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 1.2 | 3.6 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.2 | 3.6 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 1.2 | 13.2 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 1.2 | 3.6 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 1.2 | 7.0 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 1.2 | 3.5 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 1.1 | 3.4 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 1.1 | 12.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 1.1 | 12.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 1.1 | 5.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 1.1 | 5.6 | GO:0061198 | fungiform papilla development(GO:0061196) fungiform papilla morphogenesis(GO:0061197) fungiform papilla formation(GO:0061198) |
| 1.1 | 10.1 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 1.1 | 28.7 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 1.1 | 9.9 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 1.1 | 7.7 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 1.1 | 4.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 1.1 | 6.5 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.1 | 7.6 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 1.1 | 4.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.1 | 14.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 1.1 | 20.6 | GO:0042119 | neutrophil activation(GO:0042119) |
| 1.1 | 1.1 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 1.0 | 6.2 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 1.0 | 12.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 1.0 | 7.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 1.0 | 7.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 1.0 | 4.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 1.0 | 8.0 | GO:0015074 | DNA integration(GO:0015074) |
| 1.0 | 10.0 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 1.0 | 2.0 | GO:0009838 | abscission(GO:0009838) |
| 1.0 | 4.9 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 1.0 | 7.9 | GO:1902065 | response to L-glutamate(GO:1902065) |
| 1.0 | 3.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 1.0 | 8.6 | GO:1990403 | embryonic brain development(GO:1990403) |
| 1.0 | 3.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.9 | 38.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.9 | 9.3 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.9 | 5.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.9 | 9.1 | GO:1900086 | regulation of peptidyl-tyrosine autophosphorylation(GO:1900084) positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.9 | 5.4 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.9 | 4.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.9 | 24.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.9 | 1.7 | GO:0045005 | DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.8 | 18.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.8 | 6.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.8 | 19.6 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.8 | 4.9 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.8 | 2.4 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.8 | 4.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.8 | 7.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.8 | 0.8 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) |
| 0.8 | 6.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.8 | 3.1 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.8 | 3.8 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.8 | 3.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.7 | 13.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.7 | 11.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.7 | 15.6 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.7 | 2.9 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.7 | 5.2 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 0.7 | 4.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.7 | 16.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.7 | 2.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.7 | 8.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.7 | 2.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.7 | 17.3 | GO:0044783 | mitotic G1 DNA damage checkpoint(GO:0031571) G1 DNA damage checkpoint(GO:0044783) |
| 0.7 | 3.6 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.7 | 2.8 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.7 | 9.0 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.7 | 2.1 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.7 | 6.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.7 | 3.4 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.7 | 2.0 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.7 | 3.3 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.7 | 5.2 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.6 | 1.9 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.6 | 6.5 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.6 | 4.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.6 | 12.0 | GO:0007099 | centriole replication(GO:0007099) |
| 0.6 | 1.9 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.6 | 11.1 | GO:2000641 | regulation of early endosome to late endosome transport(GO:2000641) |
| 0.6 | 8.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.6 | 9.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.6 | 5.4 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.6 | 5.4 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.6 | 3.0 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.6 | 8.9 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.6 | 11.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.6 | 6.4 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.6 | 8.7 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.6 | 11.0 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.6 | 8.5 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.6 | 3.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.6 | 2.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.6 | 5.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.6 | 24.7 | GO:0017145 | stem cell division(GO:0017145) |
| 0.6 | 1.7 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.6 | 5.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.6 | 2.2 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 0.5 | 2.2 | GO:0071105 | response to interleukin-11(GO:0071105) osteoclast fusion(GO:0072675) |
| 0.5 | 5.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.5 | 3.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.5 | 2.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.5 | 11.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.5 | 2.7 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.5 | 3.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.5 | 12.1 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.5 | 1.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.5 | 1.6 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.5 | 4.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.5 | 7.5 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.5 | 6.0 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.5 | 12.9 | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition(GO:0045841) negative regulation of metaphase/anaphase transition of cell cycle(GO:1902100) |
| 0.5 | 15.8 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.5 | 8.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.5 | 5.8 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.5 | 3.9 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.5 | 12.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.5 | 1.9 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.5 | 4.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.5 | 1.8 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.5 | 0.5 | GO:0035844 | positive regulation of polarized epithelial cell differentiation(GO:0030862) cloaca development(GO:0035844) |
| 0.5 | 8.1 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.4 | 1.3 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 20.7 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.4 | 15.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.4 | 0.9 | GO:2000484 | positive regulation of interleukin-8 secretion(GO:2000484) |
| 0.4 | 1.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.4 | 10.5 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.4 | 10.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.4 | 4.7 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.4 | 4.6 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.4 | 4.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.4 | 0.4 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.4 | 3.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.4 | 3.2 | GO:0039529 | RIG-I signaling pathway(GO:0039529) |
| 0.4 | 6.7 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.4 | 6.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.4 | 8.3 | GO:0060746 | parental behavior(GO:0060746) |
| 0.4 | 6.7 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.4 | 4.3 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.4 | 2.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.4 | 2.3 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.4 | 7.6 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.4 | 5.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.4 | 6.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.4 | 3.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.4 | 2.6 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.4 | 1.8 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.4 | 1.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.4 | 2.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.4 | 1.8 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.4 | 3.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.4 | 1.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.4 | 2.8 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.4 | 3.2 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.4 | 1.4 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.4 | 6.0 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.3 | 5.2 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.3 | 37.0 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.3 | 10.6 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.3 | 1.0 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.3 | 16.8 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.3 | 23.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.3 | 3.9 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.3 | 2.9 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.3 | 4.9 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.3 | 1.6 | GO:2000973 | regulation of pro-B cell differentiation(GO:2000973) |
| 0.3 | 3.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.3 | 1.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 3.4 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.3 | 0.9 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.3 | 0.6 | GO:1904798 | regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.3 | 1.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.3 | 0.6 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.3 | 11.4 | GO:0007492 | endoderm development(GO:0007492) |
| 0.3 | 0.6 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.3 | 7.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.3 | 4.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.3 | 1.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.3 | 4.3 | GO:0032366 | intracellular lipid transport(GO:0032365) intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.3 | 0.6 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.3 | 14.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.3 | 1.7 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 4.0 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.3 | 0.5 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.3 | 6.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.3 | 1.6 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.3 | 1.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.3 | 0.8 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 8.1 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.3 | 3.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.3 | 8.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.3 | 2.0 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.3 | 1.8 | GO:0010869 | regulation of receptor biosynthetic process(GO:0010869) |
| 0.2 | 2.0 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 1.5 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.2 | 2.6 | GO:0006366 | transcription from RNA polymerase II promoter(GO:0006366) |
| 0.2 | 30.9 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.2 | 2.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 8.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.2 | 4.0 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.2 | 3.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 9.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.2 | 0.5 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.2 | 2.9 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.2 | 5.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.2 | 0.9 | GO:1902592 | viral budding(GO:0046755) multivesicular body sorting pathway(GO:0071985) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.2 | 1.3 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.2 | 1.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) response to pheromone(GO:0019236) |
| 0.2 | 6.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.2 | 5.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 6.9 | GO:0048661 | positive regulation of smooth muscle cell proliferation(GO:0048661) |
| 0.2 | 4.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 2.9 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.2 | 3.1 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 0.2 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.2 | 1.9 | GO:0035148 | embryonic epithelial tube formation(GO:0001838) neural tube formation(GO:0001841) tube formation(GO:0035148) epithelial tube formation(GO:0072175) |
| 0.2 | 0.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 4.1 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.2 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 10.9 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.2 | 1.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.2 | 1.8 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.2 | 4.2 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.2 | 2.0 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.2 | 0.5 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.2 | 11.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.2 | 48.4 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.2 | 3.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.2 | 2.8 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 2.7 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.2 | 1.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.2 | 3.5 | GO:0003170 | heart valve development(GO:0003170) |
| 0.2 | 0.7 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.2 | 1.0 | GO:0048863 | stem cell differentiation(GO:0048863) |
| 0.2 | 11.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.2 | 2.1 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 3.9 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.2 | 0.3 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.2 | 0.6 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.2 | 0.9 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 0.5 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.2 | 4.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.2 | 4.0 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 4.0 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 1.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.7 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.1 | 1.6 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 2.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 2.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 2.0 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 0.7 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 1.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.1 | 0.2 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) complement-dependent cytotoxicity(GO:0097278) |
| 0.1 | 1.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.1 | 0.8 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.7 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 2.9 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 2.4 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.1 | 0.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.6 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 3.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 7.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 1.9 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.1 | 0.9 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 2.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 9.2 | GO:0007276 | gamete generation(GO:0007276) |
| 0.1 | 0.2 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 1.3 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 7.7 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.1 | 0.2 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 | 0.2 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.1 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 5.7 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.1 | 1.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 1.7 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 1.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.6 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 2.3 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 1.9 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 1.2 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 0.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.4 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 0.6 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.1 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.4 | GO:0043278 | response to morphine(GO:0043278) |
| 0.0 | 0.4 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 1.1 | GO:0001934 | positive regulation of protein phosphorylation(GO:0001934) |
| 0.0 | 3.9 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 1.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.9 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 1.9 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.6 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.4 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.8 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.9 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.6 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 6.8 | GO:0030334 | regulation of cell migration(GO:0030334) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 | 0.7 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0045860 | positive regulation of protein kinase activity(GO:0045860) |
| 0.0 | 0.8 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 1.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.8 | 23.4 | GO:0036125 | mitochondrial fatty acid beta-oxidation multienzyme complex(GO:0016507) fatty acid beta-oxidation multienzyme complex(GO:0036125) |
| 7.5 | 29.9 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 6.1 | 61.1 | GO:0000796 | condensin complex(GO:0000796) |
| 5.1 | 30.8 | GO:0098536 | deuterosome(GO:0098536) |
| 5.0 | 20.0 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 4.8 | 19.3 | GO:0044307 | dendritic branch(GO:0044307) |
| 4.3 | 8.6 | GO:0042585 | germinal vesicle(GO:0042585) |
| 4.2 | 12.7 | GO:0005940 | septin ring(GO:0005940) |
| 3.8 | 26.8 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 3.5 | 17.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 3.3 | 9.8 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 3.3 | 13.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 3.2 | 16.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 3.0 | 11.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 2.9 | 8.8 | GO:1990047 | spindle matrix(GO:1990047) |
| 2.6 | 21.2 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 2.6 | 10.4 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 2.6 | 12.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 2.6 | 25.5 | GO:0000805 | X chromosome(GO:0000805) |
| 2.5 | 12.7 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 2.5 | 7.6 | GO:0035061 | interchromatin granule(GO:0035061) |
| 2.3 | 11.6 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 2.2 | 8.8 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 2.1 | 2.1 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 2.1 | 14.9 | GO:0001940 | male pronucleus(GO:0001940) |
| 2.1 | 12.5 | GO:0097227 | sperm annulus(GO:0097227) |
| 2.1 | 18.8 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 2.0 | 8.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 2.0 | 18.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 2.0 | 17.9 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 1.9 | 39.0 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 1.8 | 7.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 1.8 | 5.4 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 1.8 | 30.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 1.7 | 13.9 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 1.7 | 7.0 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 1.7 | 19.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 1.7 | 10.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 1.7 | 11.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.6 | 9.7 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 1.6 | 9.4 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 1.5 | 12.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.5 | 12.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.5 | 15.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.5 | 4.5 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 1.4 | 18.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 1.4 | 14.4 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 1.4 | 28.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 1.4 | 4.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 1.4 | 1.4 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 1.4 | 4.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 1.3 | 10.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 1.3 | 7.8 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 1.3 | 7.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 1.3 | 3.8 | GO:0001939 | female pronucleus(GO:0001939) |
| 1.3 | 11.5 | GO:0002177 | manchette(GO:0002177) |
| 1.3 | 2.6 | GO:0070820 | tertiary granule(GO:0070820) |
| 1.3 | 3.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 1.2 | 10.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 1.2 | 5.8 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.1 | 4.6 | GO:0032021 | NELF complex(GO:0032021) |
| 1.1 | 11.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 1.1 | 5.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 1.1 | 6.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 1.1 | 31.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 1.0 | 20.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 1.0 | 1.0 | GO:0071942 | XPC complex(GO:0071942) |
| 1.0 | 8.2 | GO:0031415 | NatA complex(GO:0031415) |
| 1.0 | 7.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 1.0 | 3.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 1.0 | 11.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 1.0 | 21.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 1.0 | 19.8 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 1.0 | 7.8 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 1.0 | 1.9 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.9 | 6.5 | GO:0070187 | telosome(GO:0070187) |
| 0.9 | 4.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.9 | 4.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.9 | 11.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.8 | 8.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.8 | 8.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.8 | 5.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.8 | 44.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.8 | 7.9 | GO:0001741 | XY body(GO:0001741) |
| 0.8 | 26.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.8 | 3.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.8 | 9.8 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.8 | 6.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.7 | 5.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.7 | 3.7 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.7 | 3.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.7 | 5.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.7 | 7.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.6 | 1.9 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.6 | 1.9 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.6 | 11.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.6 | 6.7 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.6 | 5.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.6 | 10.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.6 | 4.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 2.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.6 | 7.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.6 | 4.6 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.6 | 8.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.5 | 1.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.5 | 8.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.5 | 10.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.5 | 40.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.5 | 7.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.5 | 3.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.5 | 3.3 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.5 | 5.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.5 | 6.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.5 | 16.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.5 | 10.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.5 | 6.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 2.7 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.4 | 1.8 | GO:0001652 | granular component(GO:0001652) |
| 0.4 | 19.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.4 | 7.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.4 | 4.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.4 | 4.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 7.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.4 | 2.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.4 | 24.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.4 | 3.1 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.4 | 3.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.3 | 11.8 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.3 | 10.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 51.8 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.3 | 10.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 1.7 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.3 | 5.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 10.4 | GO:0000791 | euchromatin(GO:0000791) |
| 0.3 | 4.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 8.8 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.3 | 4.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.3 | 2.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 16.0 | GO:0005814 | centriole(GO:0005814) |
| 0.3 | 8.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.3 | 1.2 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.3 | 1.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.3 | 3.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 22.8 | GO:0016605 | PML body(GO:0016605) |
| 0.3 | 1.7 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 1.9 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.3 | 2.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 15.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.3 | 24.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.3 | 2.2 | GO:0034702 | ion channel complex(GO:0034702) |
| 0.3 | 13.6 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.2 | 9.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 1.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.4 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.2 | 2.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 13.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.2 | 3.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 74.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.2 | 3.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 5.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.2 | 11.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 0.6 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 1.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 15.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.2 | 11.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 1.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 2.8 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.2 | 83.6 | GO:0016604 | nuclear body(GO:0016604) |
| 0.2 | 1.0 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.2 | 6.6 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 33.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.2 | 1.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 6.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 6.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 2.6 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.2 | 1.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.2 | 6.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 3.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.2 | 1.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.2 | 8.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 33.9 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.2 | 3.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 2.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.2 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 18.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.7 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.1 | 0.8 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 9.8 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.1 | 8.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 20.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 10.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 3.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 3.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 5.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 3.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 1.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 4.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 16.7 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 14.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 7.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 11.0 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.1 | 3.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 10.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 3.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 12.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 5.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 6.5 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 6.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 87.0 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.1 | 1.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 4.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 2.4 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 7.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 13.2 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 1.2 | GO:0043292 | contractile fiber(GO:0043292) |
| 0.0 | 0.7 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 5.5 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 55.8 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 7.4 | 22.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 6.1 | 18.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 5.9 | 23.4 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 4.9 | 34.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 4.8 | 33.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 4.6 | 13.9 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 4.5 | 26.8 | GO:0097016 | L27 domain binding(GO:0097016) |
| 4.3 | 29.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 4.1 | 16.5 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 4.0 | 20.0 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 3.6 | 25.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 3.5 | 10.6 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 3.3 | 16.7 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 3.1 | 9.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 2.9 | 11.6 | GO:0036033 | mediator complex binding(GO:0036033) |
| 2.9 | 17.4 | GO:0097001 | ceramide binding(GO:0097001) |
| 2.7 | 13.7 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 2.6 | 33.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 2.5 | 7.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 2.4 | 7.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 2.3 | 7.0 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 2.3 | 2.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 2.1 | 8.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 2.0 | 14.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 2.0 | 20.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 1.9 | 15.6 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 1.9 | 7.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.9 | 15.0 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 1.9 | 5.6 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 1.9 | 11.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 1.8 | 12.9 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 1.8 | 20.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 1.8 | 11.0 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 1.8 | 3.6 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 1.8 | 5.4 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 1.7 | 38.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 1.7 | 6.8 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 1.5 | 7.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 1.5 | 10.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 1.5 | 4.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 1.5 | 7.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 1.4 | 14.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 1.4 | 6.8 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 1.4 | 13.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 1.4 | 8.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 1.3 | 2.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 1.3 | 9.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 1.3 | 11.9 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 1.3 | 7.8 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 1.3 | 3.8 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 1.3 | 5.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 1.3 | 10.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 1.3 | 15.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.3 | 11.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 1.2 | 7.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 1.2 | 9.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.2 | 6.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 1.2 | 3.6 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 1.2 | 5.8 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.1 | 3.4 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.1 | 21.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 1.1 | 9.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 1.1 | 2.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 1.1 | 4.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 1.1 | 5.5 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 1.1 | 4.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 1.1 | 4.3 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 1.1 | 39.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 1.1 | 6.3 | GO:0070728 | leucine binding(GO:0070728) |
| 1.1 | 8.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.0 | 29.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 1.0 | 6.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 1.0 | 4.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 1.0 | 3.9 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 1.0 | 2.9 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 1.0 | 1.9 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.0 | 14.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 1.0 | 4.8 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.9 | 3.7 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.9 | 46.9 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.9 | 5.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.8 | 5.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.8 | 42.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.8 | 5.0 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.8 | 16.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.8 | 3.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.8 | 17.0 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.8 | 15.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.8 | 7.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.8 | 51.4 | GO:0034212 | peptide N-acetyltransferase activity(GO:0034212) |
| 0.8 | 1.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.8 | 13.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.8 | 8.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.7 | 24.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.7 | 13.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.7 | 4.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.7 | 2.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.7 | 3.3 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.7 | 53.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.6 | 6.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.6 | 22.9 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.6 | 30.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.6 | 3.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.6 | 1.9 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.6 | 10.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.6 | 11.4 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.6 | 24.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.6 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.6 | 12.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.6 | 12.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.6 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.6 | 16.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.6 | 1.7 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.5 | 6.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.5 | 0.5 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.5 | 10.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.5 | 12.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.5 | 13.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.5 | 1.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.5 | 4.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.5 | 15.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.5 | 7.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.5 | 9.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.5 | 4.0 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.5 | 3.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.5 | 6.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.5 | 14.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.5 | 14.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.5 | 5.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.5 | 1.8 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.4 | 10.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.4 | 12.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.4 | 21.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.4 | 14.4 | GO:0008235 | metalloexopeptidase activity(GO:0008235) |
| 0.4 | 42.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.4 | 2.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.4 | 3.8 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.4 | 3.8 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.4 | 1.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.4 | 5.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.4 | 10.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.4 | 1.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 1.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.4 | 6.7 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.4 | 1.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.4 | 7.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.4 | 9.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.4 | 29.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.4 | 4.1 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.4 | 11.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.4 | 3.6 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.4 | 3.5 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.3 | 4.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.3 | 6.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 92.6 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.3 | 3.3 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.3 | 6.7 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.3 | 1.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 2.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.3 | 6.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 4.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 57.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.3 | 11.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.3 | 5.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.3 | 13.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.3 | 1.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.3 | 20.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.3 | 8.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.3 | 119.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.3 | 3.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.3 | 1.7 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.3 | 5.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 9.7 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.2 | 2.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.2 | 3.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.2 | 5.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 7.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 5.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 3.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 43.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.2 | 3.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 5.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 2.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 90.8 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.2 | 4.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.2 | 1.0 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.2 | 3.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.2 | 3.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 1.7 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 6.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 14.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.2 | 6.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 2.0 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.2 | 2.8 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.2 | 1.6 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.2 | 2.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.2 | 10.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.2 | 0.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 0.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.2 | 2.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 2.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 0.5 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.2 | 1.7 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 4.7 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 5.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 10.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 99.6 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 3.5 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.9 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 5.0 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 1.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 3.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 2.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.0 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 11.4 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 1.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 22.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 2.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.4 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 7.6 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 3.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.0 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.9 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 54.4 | GO:0003677 | DNA binding(GO:0003677) |
| 0.1 | 2.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.2 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.1 | 1.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.9 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 2.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.1 | 1.8 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 12.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 5.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 5.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 0.2 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 2.4 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 3.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.1 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 2.9 | GO:0005525 | GTP binding(GO:0005525) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 107.9 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 1.4 | 19.4 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 1.0 | 7.3 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 1.0 | 61.5 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.9 | 20.4 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.9 | 21.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.8 | 34.9 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.7 | 30.3 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.7 | 27.5 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.7 | 18.2 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.7 | 21.9 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.7 | 60.0 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.7 | 25.1 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 0.7 | 17.0 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 0.6 | 5.7 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.6 | 9.0 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.5 | 28.7 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.4 | 16.7 | PID_ATM_PATHWAY | ATM pathway |
| 0.4 | 18.3 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.4 | 4.5 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.4 | 15.0 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.4 | 2.9 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.3 | 6.2 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.3 | 7.7 | PID_NFAT_TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.3 | 6.1 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.3 | 7.1 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.2 | 5.9 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.2 | 4.4 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 1.2 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.2 | 5.8 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.2 | 10.7 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.2 | 3.1 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 4.1 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.1 | 5.7 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.1 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 4.3 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.1 | 1.8 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 2.3 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.5 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.6 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 4.6 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.3 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.1 | 1.9 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 17.2 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 4.7 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.6 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.5 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.3 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 1.8 | PID_SMAD2_3NUCLEAR_PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.1 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.7 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.6 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 4.0 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 3.2 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 1.2 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.3 | 8.6 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 2.9 | 2.9 | REACTOME_THROMBOXANE_SIGNALLING_THROUGH_TP_RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 2.7 | 73.0 | REACTOME_KINESINS | Genes involved in Kinesins |
| 1.8 | 99.5 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 1.7 | 29.7 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.6 | 47.8 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 1.5 | 21.1 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.4 | 24.2 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 1.3 | 69.7 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 1.3 | 54.8 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 1.3 | 7.5 | REACTOME_IRAK2_MEDIATED_ACTIVATION_OF_TAK1_COMPLEX_UPON_TLR7_8_OR_9_STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 1.2 | 4.7 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 1.1 | 10.1 | REACTOME_REGULATION_OF_THE_FANCONI_ANEMIA_PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.0 | 31.9 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.9 | 10.9 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.8 | 5.9 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.8 | 11.7 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.8 | 34.1 | REACTOME_RNA_POL_III_TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.8 | 14.5 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.7 | 28.6 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.7 | 17.6 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.6 | 6.2 | REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.6 | 13.6 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.6 | 10.1 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.5 | 15.9 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.5 | 7.0 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.5 | 9.6 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.5 | 8.5 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 46.3 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.5 | 36.2 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.5 | 14.9 | REACTOME_TRANSPORT_OF_RIBONUCLEOPROTEINS_INTO_THE_HOST_NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.4 | 8.5 | REACTOME_TRANSCRIPTIONAL_ACTIVITY_OF_SMAD2_SMAD3_SMAD4_HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.4 | 9.4 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.4 | 3.5 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.4 | 6.9 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.4 | 12.4 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.4 | 5.2 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 9.3 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.4 | 4.6 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 14.1 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.3 | 4.8 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.3 | 18.1 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.3 | 1.9 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 2.6 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.3 | 7.1 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.3 | 6.4 | REACTOME_TRAF6_MEDIATED_NFKB_ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.3 | 9.8 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.3 | 4.0 | REACTOME_NUCLEAR_EVENTS_KINASE_AND_TRANSCRIPTION_FACTOR_ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.3 | 3.3 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.3 | 7.7 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.3 | 1.6 | REACTOME_RNA_POL_II_PRE_TRANSCRIPTION_EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.3 | 0.8 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 8.2 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 7.5 | REACTOME_PEROXISOMAL_LIPID_METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 1.9 | REACTOME_PROCESSING_OF_CAPPED_INTRON_CONTAINING_PRE_MRNA | Genes involved in Processing of Capped Intron-Containing Pre-mRNA |
| 0.2 | 7.6 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 12.7 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 2.8 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 3.1 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 5.8 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 13.1 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 6.0 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 3.8 | REACTOME_PRE_NOTCH_EXPRESSION_AND_PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.2 | 6.5 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 2.5 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.2 | 4.3 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 2.8 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 2.6 | REACTOME_PIP3_ACTIVATES_AKT_SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 6.5 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 2.4 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 4.0 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.0 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.1 | REACTOME_THE_ACTIVATION_OF_ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.5 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.0 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.0 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 3.2 | REACTOME_SPHINGOLIPID_METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 8.6 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.9 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.4 | REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.1 | REACTOME_CD28_CO_STIMULATION | Genes involved in CD28 co-stimulation |
| 0.1 | 5.4 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.7 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 1.5 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.5 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.1 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.4 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.6 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.6 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 2.1 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.6 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.1 | 0.6 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 2.6 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.3 | REACTOME_DNA_REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.1 | REACTOME_GRB2_SOS_PROVIDES_LINKAGE_TO_MAPK_SIGNALING_FOR_INTERGRINS_ | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 3.1 | REACTOME_GLUCOSE_METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.5 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.9 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME_SEMA3A_PAK_DEPENDENT_AXON_REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 4.7 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |