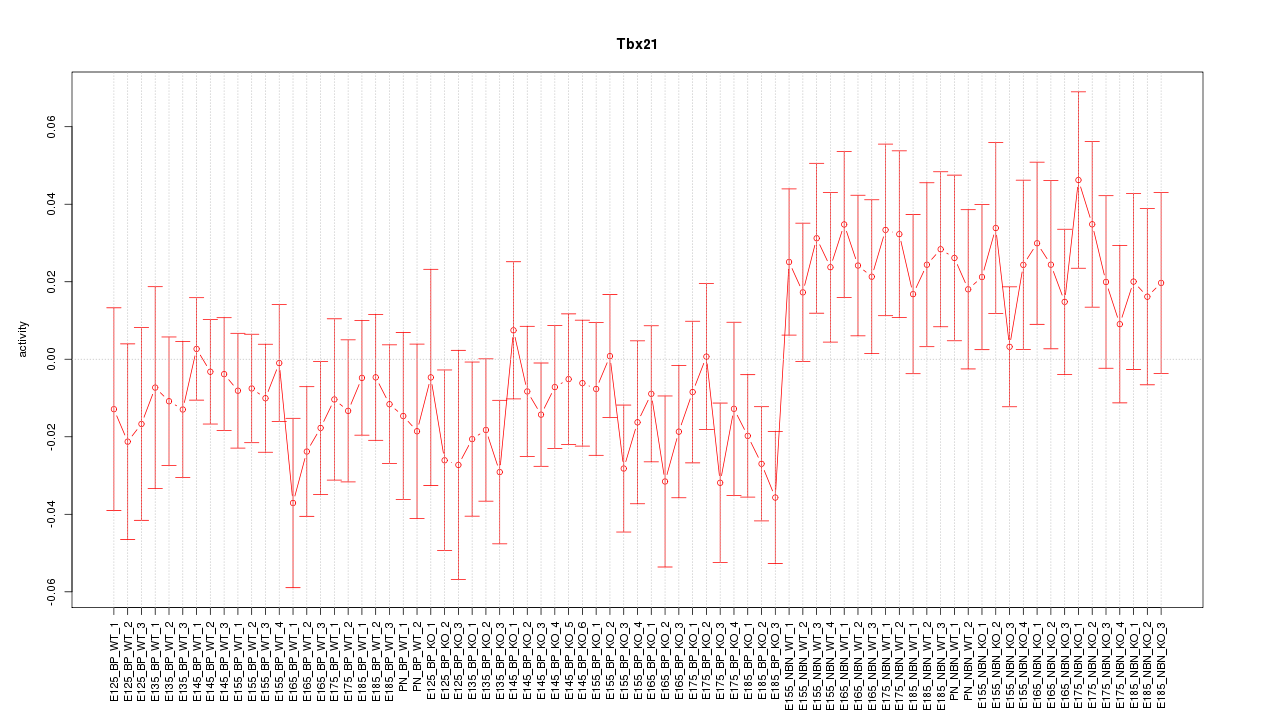
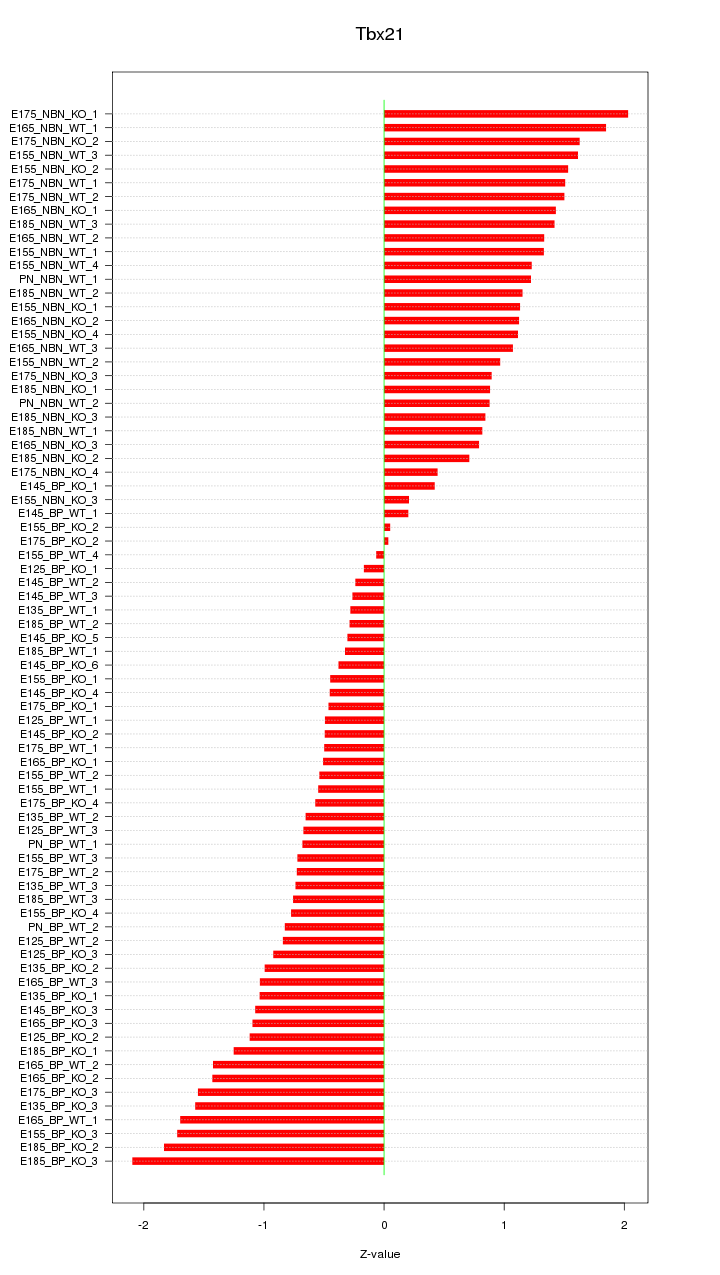
Motif ID: Tbx21
Z-value: 1.040

Transcription factors associated with Tbx21:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tbx21 | ENSMUSG00000001444.2 | Tbx21 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 16.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 2.3 | 37.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.9 | 7.5 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.6 | 6.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 1.4 | 4.3 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 1.2 | 3.5 | GO:0010248 | B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 1.0 | 3.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 1.0 | 3.0 | GO:0043379 | rRNA export from nucleus(GO:0006407) memory T cell differentiation(GO:0043379) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 1.0 | 3.9 | GO:0010040 | response to iron(II) ion(GO:0010040) positive regulation of hydrogen peroxide metabolic process(GO:0010726) negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) regulation of peroxidase activity(GO:2000468) |
| 1.0 | 2.9 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.9 | 2.7 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.8 | 4.2 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.8 | 3.2 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) |
| 0.8 | 4.9 | GO:0032796 | uropod organization(GO:0032796) |
| 0.7 | 3.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.6 | 7.7 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.6 | 34.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.5 | 2.7 | GO:0060161 | histone H4-R3 methylation(GO:0043985) positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.5 | 4.6 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.5 | 2.9 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.5 | 4.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.4 | 4.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 2.5 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.3 | 3.0 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.3 | 2.8 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 2.8 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) |
| 0.3 | 1.5 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.3 | 1.5 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.2 | 1.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 0.7 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.2 | 2.0 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.2 | 0.6 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.2 | 1.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.5 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.4 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 5.4 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 0.4 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 3.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 8.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 8.5 | GO:0060113 | inner ear receptor cell differentiation(GO:0060113) |
| 0.1 | 1.9 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.1 | 0.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 2.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 4.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 5.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 14.4 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 3.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.9 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 2.1 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 1.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 2.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.4 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.1 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.6 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 4.3 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 3.8 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0031960 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.0 | 3.3 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 3.5 | GO:0008202 | steroid metabolic process(GO:0008202) |
| 0.0 | 0.8 | GO:0006886 | intracellular protein transport(GO:0006886) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 2.4 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 2.1 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 1.0 | 3.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.6 | 2.9 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 4.9 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.4 | 2.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.4 | 6.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.4 | 2.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.4 | 1.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.4 | 3.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.4 | 37.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.3 | 3.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 3.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 14.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 2.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 2.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 2.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 7.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 4.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 7.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 7.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 3.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.9 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 4.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 5.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 3.0 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 4.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 2.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 4.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 7.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 6.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 3.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 3.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 1.6 | 6.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 1.4 | 4.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.1 | 5.4 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.7 | 3.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.7 | 2.7 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.7 | 2.7 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.7 | 3.9 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.6 | 2.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.6 | 2.9 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.6 | 14.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.5 | 1.8 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 4.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 7.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.3 | 5.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 1.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.2 | 1.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.2 | 2.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 5.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 2.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 4.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.5 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 33.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 3.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.0 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 4.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.6 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 0.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 5.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.4 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 4.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 8.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 4.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 4.8 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 7.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 3.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 15.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 26.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 3.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 2.1 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 3.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 3.5 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) C-X-C chemokine binding(GO:0019958) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.0 | 0.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 14.8 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.2 | 3.0 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 3.5 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.9 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.1 | 4.2 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.9 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.1 | 3.7 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 3.0 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 2.7 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 3.2 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID_RHOA_PATHWAY | RhoA signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 14.8 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.5 | 3.5 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 14.4 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 5.4 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 7.5 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 6.3 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 3.4 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 3.0 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 4.2 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 7.2 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.8 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.4 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 7.5 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.5 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 2.1 | REACTOME_RNA_POL_I_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 5.7 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.8 | REACTOME_METABOLISM_OF_NON_CODING_RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 0.6 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.1 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.5 | REACTOME_TRANSPORT_OF_MATURE_TRANSCRIPT_TO_CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.6 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.8 | REACTOME_RNA_POL_I_RNA_POL_III_AND_MITOCHONDRIAL_TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 3.4 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.7 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |