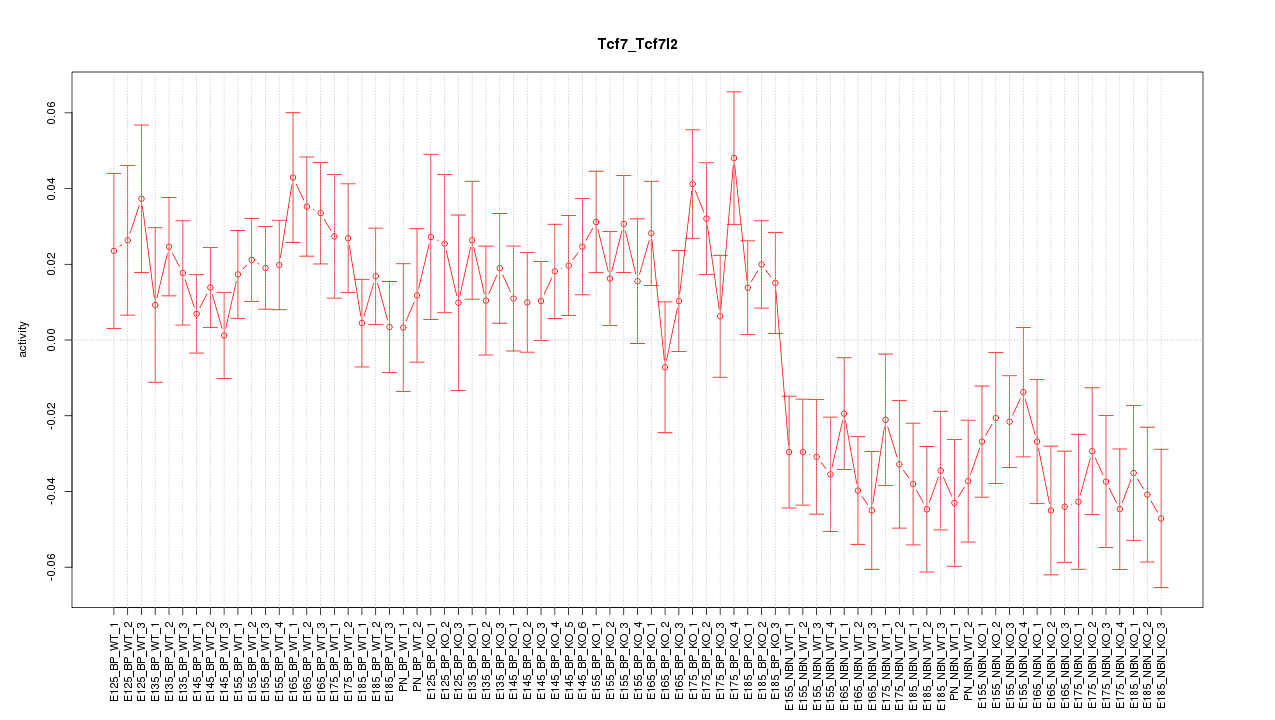
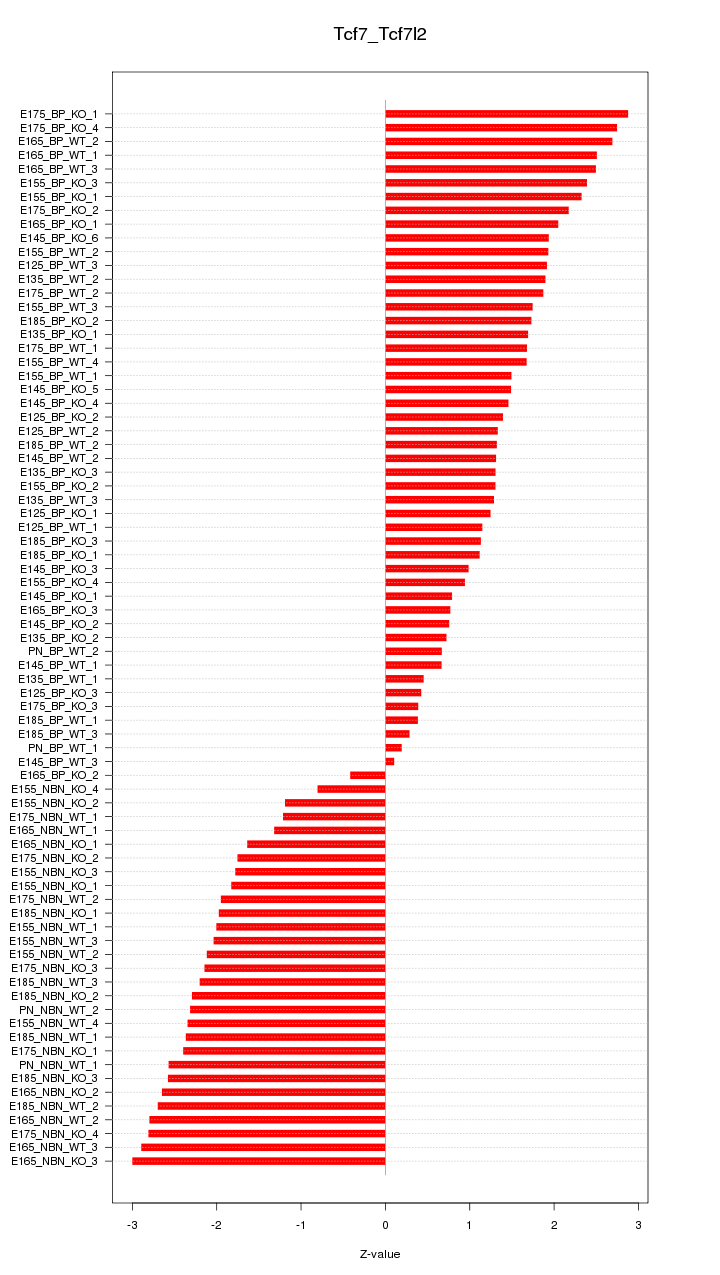
Motif ID: Tcf7_Tcf7l2
Z-value: 1.817

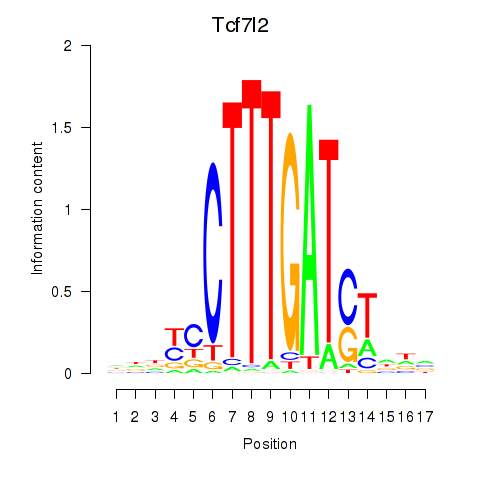
Transcription factors associated with Tcf7_Tcf7l2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tcf7 | ENSMUSG00000000782.9 | Tcf7 |
| Tcf7l2 | ENSMUSG00000024985.12 | Tcf7l2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf7 | mm10_v2_chr11_-_52282564_52282579 | -0.58 | 3.6e-08 | Click! |
| Tcf7l2 | mm10_v2_chr19_+_55742242_55742468 | 0.53 | 7.1e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 15.7 | GO:0030421 | defecation(GO:0030421) |
| 4.6 | 13.9 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 4.5 | 13.5 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 3.7 | 14.7 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 3.6 | 10.8 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 3.3 | 13.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) cellular response to lipid hydroperoxide(GO:0071449) |
| 3.3 | 9.8 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 3.1 | 21.9 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 2.6 | 7.7 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 2.1 | 10.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 1.8 | 7.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 1.8 | 7.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 1.7 | 6.6 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 1.5 | 10.7 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 1.5 | 9.1 | GO:0003383 | apical constriction(GO:0003383) |
| 1.5 | 4.5 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 1.5 | 6.0 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 1.5 | 22.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 1.3 | 8.0 | GO:0032196 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) transposition(GO:0032196) |
| 1.2 | 7.1 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 1.2 | 8.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 1.1 | 3.4 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 1.1 | 4.4 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 1.1 | 3.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 1.1 | 3.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 1.0 | 3.1 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 1.0 | 17.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 1.0 | 18.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 1.0 | 10.8 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.9 | 10.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.9 | 1.7 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.9 | 9.5 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.9 | 6.0 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.8 | 2.3 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.8 | 5.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.8 | 3.8 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.8 | 7.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.7 | 1.5 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.7 | 16.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.7 | 4.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.7 | 6.8 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.7 | 3.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.7 | 11.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.6 | 1.9 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.6 | 2.4 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.6 | 7.8 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.6 | 3.5 | GO:0060666 | pulmonary myocardium development(GO:0003350) dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.6 | 3.4 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.5 | 5.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.5 | 1.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.5 | 2.5 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.5 | 26.3 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.5 | 1.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.5 | 1.4 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.5 | 6.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.5 | 10.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.4 | 3.0 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.4 | 2.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.4 | 6.8 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.4 | 2.5 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.4 | 5.4 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.4 | 2.5 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.4 | 2.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 2.8 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.4 | 3.2 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.4 | 2.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.4 | 1.1 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.4 | 1.1 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.4 | 1.9 | GO:0007403 | blastocyst hatching(GO:0001835) glial cell fate determination(GO:0007403) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.4 | 12.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.4 | 2.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.4 | 2.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.4 | 2.5 | GO:0051461 | protein import into peroxisome matrix, docking(GO:0016560) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.3 | 3.4 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.3 | 1.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 2.0 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.3 | 16.1 | GO:0001947 | heart looping(GO:0001947) |
| 0.3 | 4.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.3 | 2.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 1.5 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.3 | 6.1 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.3 | 1.7 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.3 | 5.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.3 | 7.3 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.3 | 1.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.3 | 1.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.3 | 2.9 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.3 | 1.1 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.3 | 7.3 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.3 | 0.8 | GO:1904798 | regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.3 | 1.8 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.3 | 0.8 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.3 | 2.3 | GO:0033233 | regulation of protein sumoylation(GO:0033233) positive regulation of protein sumoylation(GO:0033235) |
| 0.3 | 1.5 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.2 | 1.9 | GO:2000303 | regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 0.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 2.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 2.6 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.2 | 2.0 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.2 | 1.4 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 1.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.8 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.2 | 2.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 10.1 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.2 | 1.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.2 | 2.0 | GO:0033129 | positive regulation of histone phosphorylation(GO:0033129) |
| 0.2 | 0.4 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.2 | 1.1 | GO:0016557 | peroxisomal membrane transport(GO:0015919) peroxisome membrane biogenesis(GO:0016557) protein import into peroxisome membrane(GO:0045046) |
| 0.2 | 2.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.2 | 6.5 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.2 | 1.6 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 7.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.2 | 9.9 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.2 | 5.9 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 | 1.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.6 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 1.6 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 6.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 1.2 | GO:0030813 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.1 | 2.2 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) Sertoli cell development(GO:0060009) |
| 0.1 | 0.1 | GO:2000911 | positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
| 0.1 | 7.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 16.2 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.1 | 1.6 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 5.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.5 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.3 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 15.5 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 1.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 1.7 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.4 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.1 | 6.6 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 3.9 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 3.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 1.8 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 9.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 7.4 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 2.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.3 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.1 | 0.4 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 1.3 | GO:0023058 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.1 | 3.9 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 3.2 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.1 | 7.0 | GO:0048864 | stem cell development(GO:0048864) |
| 0.1 | 2.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 3.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.4 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.2 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 1.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.9 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 2.7 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 1.8 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 2.7 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 1.3 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 4.0 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 2.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 1.2 | GO:0051492 | regulation of stress fiber assembly(GO:0051492) |
| 0.0 | 2.0 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.3 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 4.2 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.6 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 2.4 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 11.0 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.6 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0061365 | intestinal cholesterol absorption(GO:0030299) positive regulation of triglyceride lipase activity(GO:0061365) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 1.6 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.3 | GO:0001934 | positive regulation of protein phosphorylation(GO:0001934) |
| 0.0 | 2.5 | GO:0016311 | dephosphorylation(GO:0016311) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 4.5 | 13.5 | GO:0071914 | prominosome(GO:0071914) |
| 2.6 | 10.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 2.3 | 9.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 2.1 | 10.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.8 | 7.0 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 1.1 | 9.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.1 | 10.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.9 | 6.5 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.7 | 15.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.6 | 9.9 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.6 | 2.5 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.6 | 21.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.5 | 3.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.5 | 2.2 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.5 | 4.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.5 | 7.3 | GO:0031105 | septin complex(GO:0031105) |
| 0.4 | 6.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.4 | 1.5 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.4 | 4.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 2.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 3.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.3 | 3.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.3 | 1.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.3 | 3.9 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.3 | 1.9 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 21.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.3 | 16.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 0.8 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.3 | 13.5 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.2 | 1.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 1.9 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.2 | 2.9 | GO:0045120 | pronucleus(GO:0045120) |
| 0.2 | 19.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 1.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 1.7 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 1.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 2.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 14.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 19.6 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 13.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 2.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 6.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.2 | 8.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.9 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 2.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 2.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 3.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 5.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 10.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 9.2 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.1 | 1.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 4.7 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 1.0 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 24.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 12.1 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.1 | 2.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 3.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 1.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 8.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 4.1 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 1.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 21.9 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 2.4 | 7.3 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 2.4 | 14.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 2.0 | 6.0 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 2.0 | 8.0 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.9 | 7.7 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 1.5 | 6.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 1.3 | 6.5 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 1.3 | 5.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 1.2 | 9.9 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 1.1 | 3.2 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) oxidoreductase activity, acting on the CH-OH group of donors, cytochrome as acceptor(GO:0016898) |
| 1.1 | 3.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 1.0 | 9.8 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 1.0 | 11.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.9 | 2.6 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.8 | 12.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.8 | 15.0 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.7 | 13.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.7 | 7.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.7 | 16.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.6 | 2.5 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.6 | 10.8 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.6 | 1.7 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.6 | 2.8 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.5 | 1.6 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.5 | 1.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.5 | 5.7 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.5 | 3.0 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.5 | 2.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.5 | 1.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.5 | 6.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 7.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.5 | 11.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.4 | 3.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.4 | 8.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.4 | 2.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.4 | 3.4 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.4 | 1.9 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.4 | 4.7 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.3 | 1.4 | GO:0043546 | molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.3 | 3.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.3 | 2.5 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.3 | 3.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 1.9 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.3 | 2.4 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.3 | 1.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 1.7 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.3 | 7.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 21.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.3 | 1.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.3 | 2.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 5.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 2.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 3.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.2 | 1.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.2 | 8.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 2.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.2 | 10.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 2.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 7.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 0.8 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 3.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 13.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.2 | 2.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 32.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 5.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.2 | 2.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 3.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 0.7 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 0.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 1.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 16.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.9 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.8 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 1.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 4.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 10.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 9.2 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 9.0 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 1.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 2.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 2.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 3.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.6 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 1.8 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 5.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 24.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 0.3 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 3.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.6 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 4.2 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.1 | 24.8 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 3.2 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 3.1 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 16.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 14.5 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 2.5 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.8 | GO:0044390 | ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 1.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 6.7 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.0 | GO:0052866 | phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 21.6 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 9.4 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 2.8 | GO:0016791 | phosphatase activity(GO:0016791) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 22.1 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.6 | 8.1 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 1.5 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.5 | 1.9 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.4 | 9.2 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.4 | 14.6 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 14.0 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 12.7 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.3 | 16.4 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.3 | 12.8 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 5.4 | PID_S1P_META_PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 18.8 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.2 | 7.3 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 13.1 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.7 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.8 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 8.4 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 7.9 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 11.5 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 4.7 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.8 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.5 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.5 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 4.3 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 0.9 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 4.4 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.5 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 1.5 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.0 | 1.4 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.1 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.4 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 9.2 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.7 | 10.0 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.6 | 11.0 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.6 | 10.2 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.6 | 18.7 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.6 | 6.2 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.5 | 18.8 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.5 | 13.1 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.4 | 6.5 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 9.1 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 9.9 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.4 | 10.7 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.4 | 11.7 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.3 | 8.1 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.3 | 3.0 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 18.6 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 11.5 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 7.1 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 2.5 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 1.5 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.3 | 9.8 | REACTOME_SMAD2_SMAD3_SMAD4_HETEROTRIMER_REGULATES_TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.3 | 10.1 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 11.1 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELLULAR_PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 10.4 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 5.1 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 1.7 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 1.3 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 6.7 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 1.4 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.5 | REACTOME_COMPLEMENT_CASCADE | Genes involved in Complement cascade |
| 0.1 | 4.0 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 6.0 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.4 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.6 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.2 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.4 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 2.2 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.2 | REACTOME_SPHINGOLIPID_METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 0.7 | REACTOME_ER_PHAGOSOME_PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 1.2 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.9 | REACTOME_TRANSPORT_OF_MATURE_MRNA_DERIVED_FROM_AN_INTRONLESS_TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 2.4 | REACTOME_EXTRACELLULAR_MATRIX_ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.1 | 3.7 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 4.9 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 2.5 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.0 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.8 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 3.3 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.5 | REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 2.4 | REACTOME_MRNA_SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME_FORMATION_OF_TRANSCRIPTION_COUPLED_NER_TC_NER_REPAIR_COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 1.7 | REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | Genes involved in Generic Transcription Pathway |