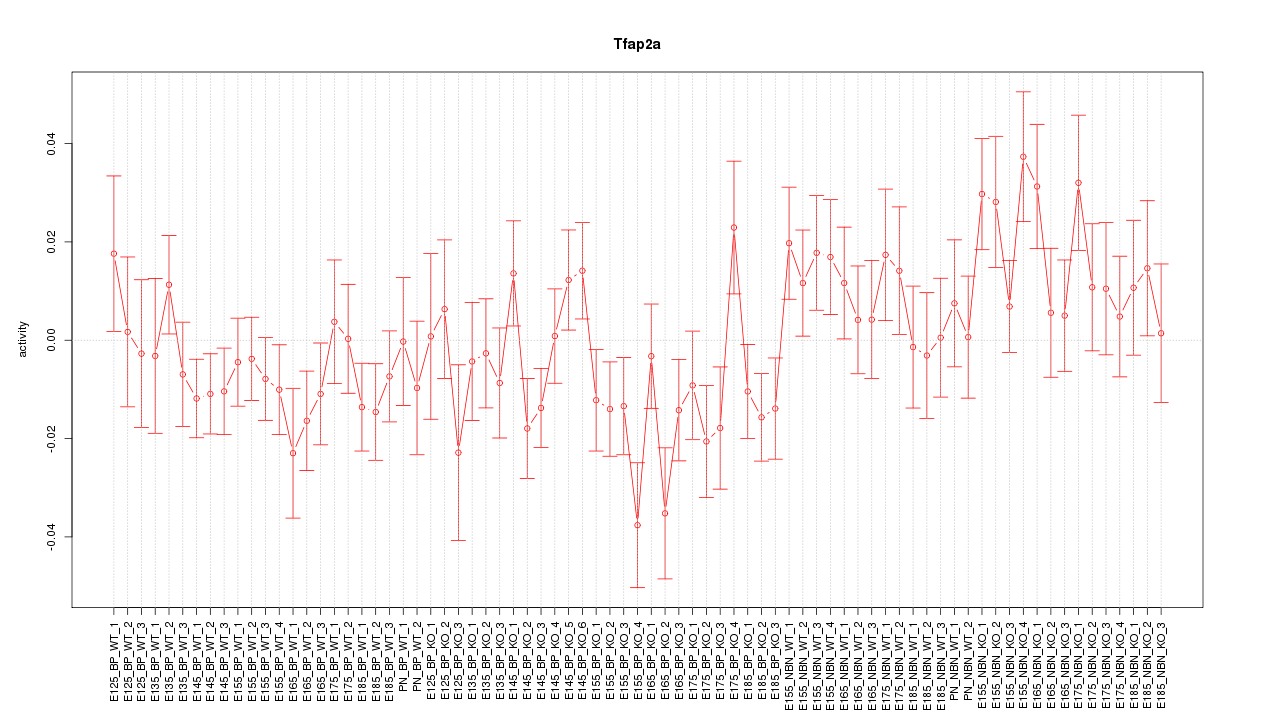
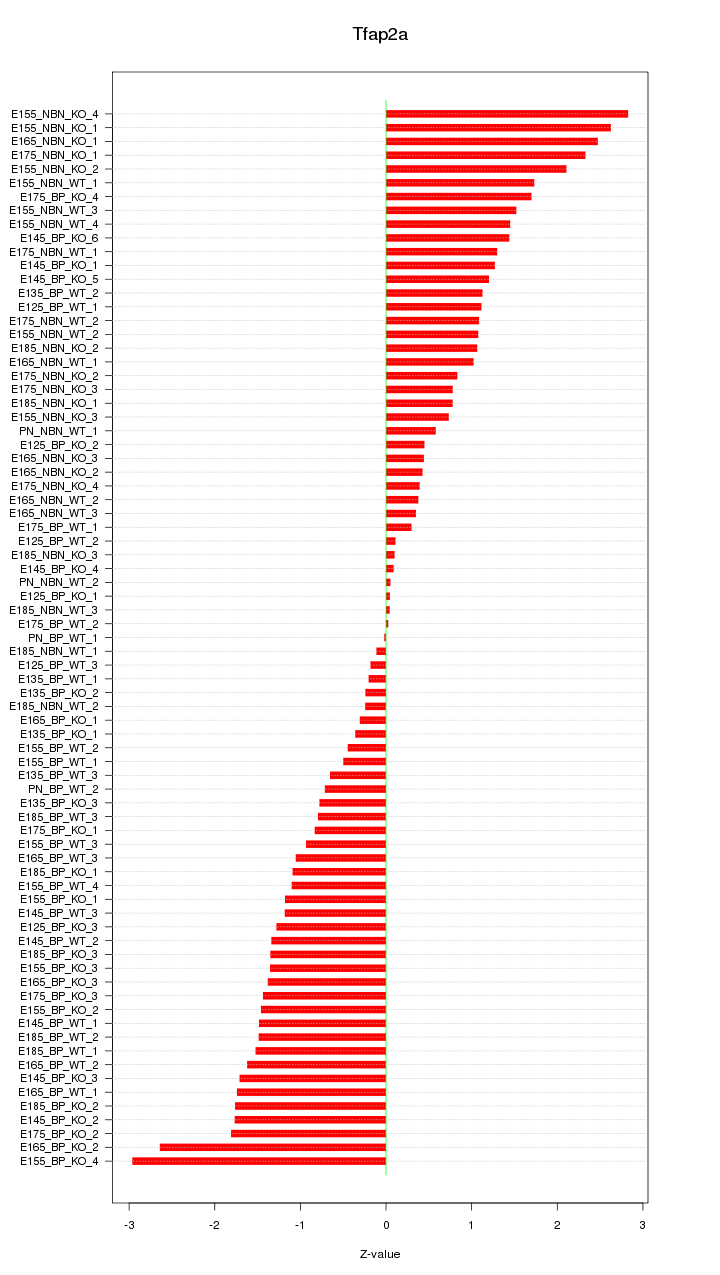
Motif ID: Tfap2a
Z-value: 1.267

Transcription factors associated with Tfap2a:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tfap2a | ENSMUSG00000021359.9 | Tfap2a |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2a | mm10_v2_chr13_-_40733768_40733836 | 0.05 | 6.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 17.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 2.5 | 10.0 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 2.3 | 7.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 2.1 | 27.3 | GO:0070842 | aggresome assembly(GO:0070842) |
| 1.9 | 7.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.7 | 5.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 1.6 | 10.9 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 1.5 | 7.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 1.4 | 7.0 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 1.2 | 4.8 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 1.2 | 3.5 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 1.1 | 4.5 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 1.1 | 22.7 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 1.1 | 6.7 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 1.1 | 3.2 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 1.0 | 3.1 | GO:0010752 | signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) |
| 1.0 | 3.0 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.9 | 7.3 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.9 | 5.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.8 | 3.4 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.8 | 2.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.8 | 3.2 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.8 | 3.9 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.7 | 8.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.7 | 2.2 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.7 | 2.8 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.7 | 7.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.7 | 2.1 | GO:0046959 | habituation(GO:0046959) |
| 0.7 | 3.4 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.7 | 2.0 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.6 | 1.9 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.6 | 2.6 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.6 | 1.2 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.6 | 2.3 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.6 | 2.9 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.6 | 2.8 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.5 | 17.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.5 | 1.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.5 | 1.6 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.5 | 2.6 | GO:0021764 | amygdala development(GO:0021764) |
| 0.5 | 2.5 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.5 | 3.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.5 | 3.5 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.5 | 1.5 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.5 | 1.5 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.5 | 5.7 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.5 | 4.6 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 3.5 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.4 | 3.1 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.4 | 2.6 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.4 | 1.7 | GO:0061047 | positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.4 | 1.2 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.4 | 1.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.4 | 3.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 2.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.4 | 2.4 | GO:0002317 | plasma cell differentiation(GO:0002317) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.4 | 3.0 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.4 | 2.2 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.4 | 6.8 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.4 | 1.4 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.4 | 1.1 | GO:0090649 | cellular response to manganese ion(GO:0071287) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.3 | 3.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 1.4 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 0.3 | 1.3 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.3 | 1.7 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 1.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.3 | 2.6 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 1.6 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.3 | 0.9 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.3 | 2.4 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.3 | 0.9 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.3 | 0.9 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.3 | 1.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.3 | 1.4 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.3 | 0.8 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.3 | 2.2 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.3 | 1.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.3 | 1.6 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.3 | 1.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.3 | 3.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.3 | 6.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 0.5 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) |
| 0.3 | 0.8 | GO:2000468 | regulation of peroxidase activity(GO:2000468) |
| 0.3 | 7.7 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.3 | 5.6 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.3 | 0.8 | GO:0032493 | response to bacterial lipoprotein(GO:0032493) |
| 0.3 | 1.0 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.2 | 0.5 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 7.0 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.2 | 3.6 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 2.6 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.2 | 1.8 | GO:0080184 | response to stilbenoid(GO:0035634) response to phenylpropanoid(GO:0080184) |
| 0.2 | 1.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 1.8 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.2 | 7.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 0.9 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.2 | 3.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.2 | 4.0 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 1.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 9.3 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.2 | 0.8 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.2 | 1.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.8 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.2 | 4.6 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 0.9 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.2 | 3.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 | 1.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.2 | 0.7 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.2 | 3.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 1.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 1.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.2 | 1.9 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.2 | 3.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.2 | 1.4 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 3.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 6.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.2 | 1.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 1.8 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.2 | 2.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 1.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 0.6 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 2.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 3.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 3.3 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.2 | 1.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.2 | 0.9 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.2 | 0.9 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.1 | 1.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 2.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 0.9 | GO:0032796 | uropod organization(GO:0032796) |
| 0.1 | 12.1 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 2.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 2.3 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.1 | 2.0 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 | 0.6 | GO:1905247 | modulation of age-related behavioral decline(GO:0090647) positive regulation of beta-amyloid clearance(GO:1900223) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) negative regulation of metalloendopeptidase activity(GO:1904684) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 0.9 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.9 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.8 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 2.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.9 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.6 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.1 | 2.7 | GO:2000772 | regulation of cellular senescence(GO:2000772) |
| 0.1 | 2.0 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 1.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 4.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.4 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.1 | 1.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 4.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.3 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 2.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 2.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.4 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 1.3 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.2 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.1 | 0.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.3 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.1 | 0.6 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 5.9 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 4.3 | GO:0030071 | regulation of mitotic metaphase/anaphase transition(GO:0030071) regulation of metaphase/anaphase transition of cell cycle(GO:1902099) |
| 0.1 | 1.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 2.0 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.1 | 3.1 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.1 | 1.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 1.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 0.3 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 1.1 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.1 | 0.9 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.2 | GO:0035087 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.1 | 0.6 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.1 | 1.7 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.1 | 0.4 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 2.0 | GO:0061157 | positive regulation of mRNA catabolic process(GO:0061014) mRNA destabilization(GO:0061157) |
| 0.1 | 0.4 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 1.2 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.1 | 0.2 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) negative regulation of intestinal absorption(GO:1904479) |
| 0.1 | 9.3 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 3.1 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 1.0 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 3.0 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 1.8 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.8 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 2.2 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 3.6 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.2 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 4.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.5 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 1.1 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.1 | 0.5 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.1 | 7.6 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.1 | 0.2 | GO:0015867 | ATP transport(GO:0015867) |
| 0.1 | 1.1 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 1.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 1.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.5 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 1.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 1.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 3.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 1.7 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 1.4 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 7.0 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 3.0 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 1.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 1.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 1.1 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 2.7 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 1.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.1 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 1.1 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.3 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.0 | 0.0 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.9 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.7 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.5 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 1.3 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 1.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.5 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.6 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.7 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 0.7 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.5 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 10.0 | GO:0072534 | perineuronal net(GO:0072534) |
| 2.3 | 7.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 2.2 | 6.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.7 | 6.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.5 | 7.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 1.2 | 3.7 | GO:0099573 | glutamatergic postsynaptic density(GO:0099573) |
| 0.9 | 3.5 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.8 | 12.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.7 | 0.7 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.7 | 2.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.6 | 7.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.6 | 1.8 | GO:0005940 | septin ring(GO:0005940) |
| 0.5 | 12.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.4 | 1.3 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.4 | 4.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.4 | 2.9 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.4 | 5.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.4 | 6.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 3.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.3 | 1.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.3 | 2.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 1.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.3 | 1.7 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 4.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 1.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 3.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 2.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.2 | 2.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 3.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 5.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 0.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 1.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 0.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 8.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 19.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.2 | 0.9 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 1.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 1.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 0.5 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.2 | 11.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 14.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 4.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 8.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 5.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 3.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.9 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 2.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.7 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 2.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 1.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 2.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 1.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 37.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 3.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 13.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 2.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.5 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 1.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 2.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 7.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 1.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 2.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 1.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 2.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.7 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.1 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 7.1 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.8 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.9 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 2.3 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 1.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 15.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.9 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 1.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 2.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 5.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 1.9 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 3.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0098533 | ATPase dependent transmembrane transport complex(GO:0098533) |
| 0.0 | 0.9 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 11.3 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.0 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 1.4 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 26.0 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.9 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 2.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 22.7 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 2.8 | 17.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 1.8 | 5.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 1.7 | 6.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 1.5 | 7.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.4 | 4.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 1.4 | 10.9 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 1.0 | 6.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.9 | 1.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.9 | 3.5 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.8 | 13.5 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.7 | 2.8 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.7 | 10.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.7 | 2.0 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.7 | 2.0 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.6 | 2.4 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.5 | 1.6 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.5 | 3.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.5 | 2.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 2.5 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.5 | 1.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.5 | 2.5 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.5 | 2.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.5 | 1.9 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.5 | 1.9 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.5 | 2.9 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.5 | 2.3 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.5 | 3.7 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.5 | 2.7 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.5 | 8.6 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.4 | 1.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.4 | 2.2 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.4 | 14.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.4 | 1.2 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.4 | 1.2 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 1.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.4 | 1.2 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.4 | 2.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.4 | 3.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.4 | 10.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.4 | 1.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.4 | 3.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.4 | 1.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.4 | 3.9 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.3 | 1.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.3 | 6.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.3 | 7.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.3 | 12.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 3.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.3 | 0.9 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.3 | 1.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.3 | 2.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 2.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 1.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 2.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 0.8 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.2 | 1.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 2.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 1.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 0.7 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) single guanine insertion binding(GO:0032142) |
| 0.2 | 3.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.2 | 1.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 4.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 7.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 3.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.2 | 1.9 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 11.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.2 | 0.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 4.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.2 | 0.7 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.2 | 0.5 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 5.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 1.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 0.8 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.2 | 3.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.2 | 2.0 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 3.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 0.9 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 0.9 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.2 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 3.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 1.1 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.1 | 1.5 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 1.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 2.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.8 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.1 | 0.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.6 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 1.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.9 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 5.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 2.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.8 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 1.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 1.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 9.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 1.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 11.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 4.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 3.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 7.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.3 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.1 | 11.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 5.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 2.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.8 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 7.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 3.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.2 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 1.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.3 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 1.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 5.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 2.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 5.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 13.5 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.8 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 5.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 3.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:1990430 | G-protein coupled GABA receptor activity(GO:0004965) extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 1.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.4 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.1 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 3.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.3 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.3 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 7.8 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 5.7 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.2 | 1.9 | PID_CONE_PATHWAY | Visual signal transduction: Cones |
| 0.2 | 4.0 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 0.9 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.0 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 3.1 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.2 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 4.6 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.3 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 4.0 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 4.3 | PID_ATM_PATHWAY | ATM pathway |
| 0.1 | 3.7 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.1 | 3.5 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.1 | 2.0 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.7 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.6 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 11.7 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.2 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 4.1 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.1 | 0.8 | PID_AVB3_OPN_PATHWAY | Osteopontin-mediated events |
| 0.1 | 3.3 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 13.0 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.4 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 8.8 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.9 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.4 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.5 | PID_VEGFR1_2_PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.3 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.8 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.0 | 2.6 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.4 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.6 | PID_A6B1_A6B4_INTEGRIN_PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.8 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.5 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.8 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.0 | 0.8 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 17.0 | REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.7 | 22.7 | REACTOME_A_TETRASACCHARIDE_LINKER_SEQUENCE_IS_REQUIRED_FOR_GAG_SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.5 | 12.5 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.5 | 13.0 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.5 | 15.2 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.4 | 2.1 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 0.4 | 8.2 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.4 | 8.6 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.4 | 5.2 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 3.4 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.3 | 5.7 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 11.6 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 0.8 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.2 | 3.0 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.2 | 7.7 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 2.5 | REACTOME_SPRY_REGULATION_OF_FGF_SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 2.8 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 2.5 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.3 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 4.2 | REACTOME_TRANSFERRIN_ENDOCYTOSIS_AND_RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.2 | 2.2 | REACTOME_DSCAM_INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.3 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.8 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 0.8 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.7 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 4.9 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.1 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.5 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.2 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.5 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.1 | 4.2 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.8 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 3.0 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.6 | REACTOME_FORMATION_OF_FIBRIN_CLOT_CLOTTING_CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 0.9 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.6 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.3 | REACTOME_RNA_POL_III_TRANSCRIPTION_TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.9 | REACTOME_PKA_MEDIATED_PHOSPHORYLATION_OF_CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.8 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.2 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.2 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.0 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.5 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.1 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 0.3 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 0.8 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 6.7 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.6 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.4 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.5 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 3.2 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.9 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.1 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.1 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 3.4 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.1 | REACTOME_RNA_POL_I_TRANSCRIPTION_INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.2 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.4 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.3 | REACTOME_SPHINGOLIPID_METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 2.2 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.2 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.0 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 2.2 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.5 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME_LIPID_DIGESTION_MOBILIZATION_AND_TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.7 | REACTOME_REGULATION_OF_GLUCOKINASE_BY_GLUCOKINASE_REGULATORY_PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 1.8 | REACTOME_TRANSMISSION_ACROSS_CHEMICAL_SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.7 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.1 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |