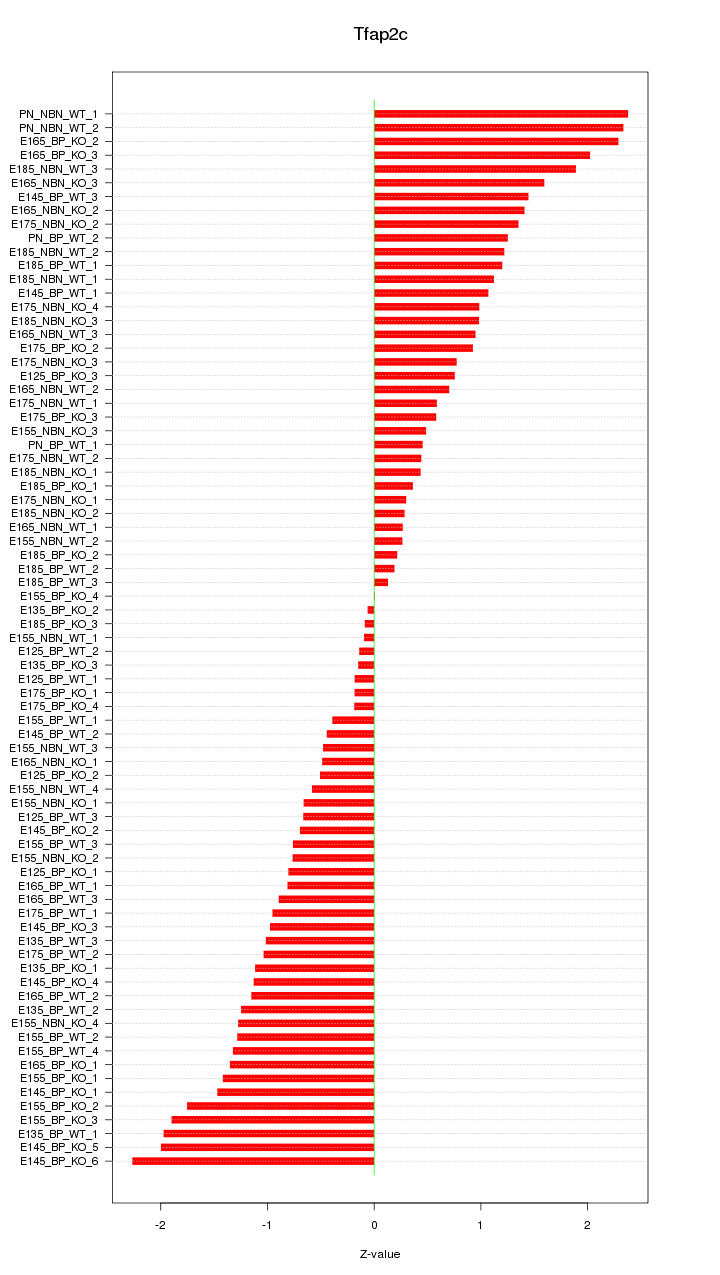
Motif ID: Tfap2c
Z-value: 1.100
Transcription factors associated with Tfap2c:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tfap2c | ENSMUSG00000028640.5 | Tfap2c |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2c | mm10_v2_chr2_+_172550761_172550782 | -0.51 | 2.0e-06 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 15.7 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 2.4 | 7.3 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 2.4 | 9.6 | GO:0050904 | diapedesis(GO:0050904) |
| 2.3 | 7.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 2.2 | 13.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 2.0 | 6.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 1.8 | 23.5 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 1.7 | 11.9 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.7 | 5.1 | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 1.6 | 7.9 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.2 | 3.5 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.1 | 3.4 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 1.1 | 4.4 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.9 | 2.7 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.8 | 4.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.8 | 3.3 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.7 | 2.9 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.7 | 2.7 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.7 | 2.7 | GO:0060032 | notochord regression(GO:0060032) |
| 0.6 | 5.0 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.6 | 4.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.6 | 1.8 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.6 | 2.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.6 | 2.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.6 | 1.8 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.6 | 2.4 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.6 | 2.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) lateral motor column neuron migration(GO:0097477) |
| 0.6 | 3.3 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.6 | 1.1 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.5 | 1.5 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.5 | 2.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.5 | 2.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.5 | 3.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.5 | 7.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.4 | 2.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.4 | 1.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.4 | 1.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.4 | 3.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 2.8 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.4 | 1.5 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) |
| 0.4 | 0.7 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.4 | 1.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.4 | 4.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.4 | 1.8 | GO:0015822 | ornithine transport(GO:0015822) |
| 0.3 | 3.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.3 | 1.4 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.3 | 3.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.3 | 3.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 1.0 | GO:0033122 | regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.3 | 1.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.3 | 1.6 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.3 | 1.6 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.3 | 0.9 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.3 | 0.9 | GO:0046104 | thymidine metabolic process(GO:0046104) |
| 0.3 | 1.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.3 | 0.8 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.3 | 1.1 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.3 | 1.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.3 | 1.9 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.3 | 2.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.3 | 1.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.3 | 1.6 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.3 | 3.6 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.3 | 2.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 3.8 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.2 | 5.4 | GO:0048535 | lymph node development(GO:0048535) |
| 0.2 | 1.4 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.2 | 0.7 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.2 | 2.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.2 | 0.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.2 | 1.6 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 | 2.0 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.2 | 2.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.7 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.2 | 2.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.2 | 0.8 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.2 | 0.8 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 1.0 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 | 5.1 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.2 | 3.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 1.2 | GO:2000348 | protein linear polyubiquitination(GO:0097039) regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 1.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.2 | 0.7 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.2 | 3.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 6.3 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.2 | 5.3 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 2.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 1.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.3 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.1 | 0.6 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 1.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.7 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.4 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.1 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 1.8 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 1.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.4 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 3.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 1.0 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.4 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 4.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 1.4 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 0.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 3.3 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.1 | 10.7 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.7 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.1 | 4.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.1 | 0.7 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.6 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 1.4 | GO:0006023 | aminoglycan biosynthetic process(GO:0006023) |
| 0.0 | 2.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.2 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 1.5 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 1.0 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 3.0 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 1.0 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 2.9 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 1.0 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.7 | GO:0046879 | hormone secretion(GO:0046879) |
| 0.0 | 1.8 | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway(GO:0007169) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.6 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 5.2 | 15.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.9 | 9.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 1.1 | 7.9 | GO:0005638 | lamin filament(GO:0005638) |
| 1.0 | 6.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.8 | 3.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.7 | 2.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.5 | 11.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.4 | 1.3 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.3 | 3.8 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 5.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.3 | 1.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 5.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 3.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 3.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 2.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.2 | 3.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.2 | 0.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 3.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 3.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 2.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 3.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 2.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 1.0 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 4.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 14.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.0 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 3.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 2.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 2.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 3.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.4 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 2.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 2.7 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.7 | GO:0043657 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.0 | 0.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 2.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 4.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.4 | GO:0071944 | cell periphery(GO:0071944) |
| 0.0 | 0.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 4.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 3.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 2.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.8 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 4.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 3.0 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 3.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 4.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 13.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 3.0 | 11.9 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 2.2 | 15.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 1.5 | 22.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.1 | 3.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.0 | 3.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.9 | 2.7 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.8 | 7.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.8 | 2.4 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.7 | 5.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.7 | 2.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.7 | 2.0 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.7 | 2.0 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.6 | 1.9 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.6 | 2.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.6 | 2.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.6 | 3.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.5 | 2.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.5 | 4.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 1.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.5 | 3.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.4 | 1.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 1.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.4 | 3.4 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.4 | 1.5 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.4 | 2.9 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.4 | 3.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 2.0 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 2.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 1.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.3 | 1.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.3 | 1.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 1.6 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.3 | 1.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.3 | 5.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.3 | 9.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.3 | 0.8 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 1.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.3 | 7.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 6.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 1.3 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 0.9 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.2 | 1.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.9 | GO:0032564 | dATP binding(GO:0032564) |
| 0.2 | 4.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 2.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.2 | 4.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 2.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.2 | 1.8 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 0.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 7.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 0.9 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.1 | 1.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.8 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 2.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 1.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 1.8 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.0 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) |
| 0.1 | 1.0 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 2.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 2.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.6 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 2.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.8 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 1.0 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.1 | 1.1 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.1 | 4.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 3.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 7.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.0 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 1.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 2.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 10.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 3.0 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 2.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.5 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 2.3 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 2.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.8 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.6 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 2.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 4.1 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 3.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 6.0 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 3.8 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 11.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 2.6 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 1.6 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.8 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 1.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.1 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 30.2 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.4 | 3.3 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.3 | 4.6 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.3 | 7.5 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.3 | 4.8 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 6.9 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.2 | 8.3 | PID_IL8_CXCR2_PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 4.4 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 1.6 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 3.4 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 6.2 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 18.3 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 1.9 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.9 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.5 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 12.3 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.2 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.2 | PID_SYNDECAN_2_PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.9 | PID_MAPK_TRK_PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.3 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.0 | 1.3 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.9 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.3 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.4 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.5 | PID_AVB3_INTEGRIN_PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 3.2 | NABA_MATRISOME_ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.5 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.7 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 1.3 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID_IL2_PI3K_PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID_LKB1_PATHWAY | LKB1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 22.8 | REACTOME_PLATELET_ADHESION_TO_EXPOSED_COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.7 | 9.6 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.7 | 7.3 | REACTOME_SIGNALING_BY_ACTIVATED_POINT_MUTANTS_OF_FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.4 | 3.2 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 3.4 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 13.0 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 11.3 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 2.2 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 3.3 | REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.3 | 3.3 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 2.4 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 2.1 | REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.2 | 4.3 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 12.7 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 1.9 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 3.0 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.6 | REACTOME_RAF_MAP_KINASE_CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.6 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.1 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.8 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.8 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 5.0 | REACTOME_THROMBIN_SIGNALLING_THROUGH_PROTEINASE_ACTIVATED_RECEPTORS_PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 2.9 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.1 | 0.9 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.0 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.8 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.3 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.5 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.7 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.1 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.3 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 1.4 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.8 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.8 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.0 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.3 | REACTOME_DAG_AND_IP3_SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 1.0 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.8 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.2 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 3.4 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 2.1 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.7 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 1.6 | REACTOME_GLYCEROPHOSPHOLIPID_BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 1.0 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |