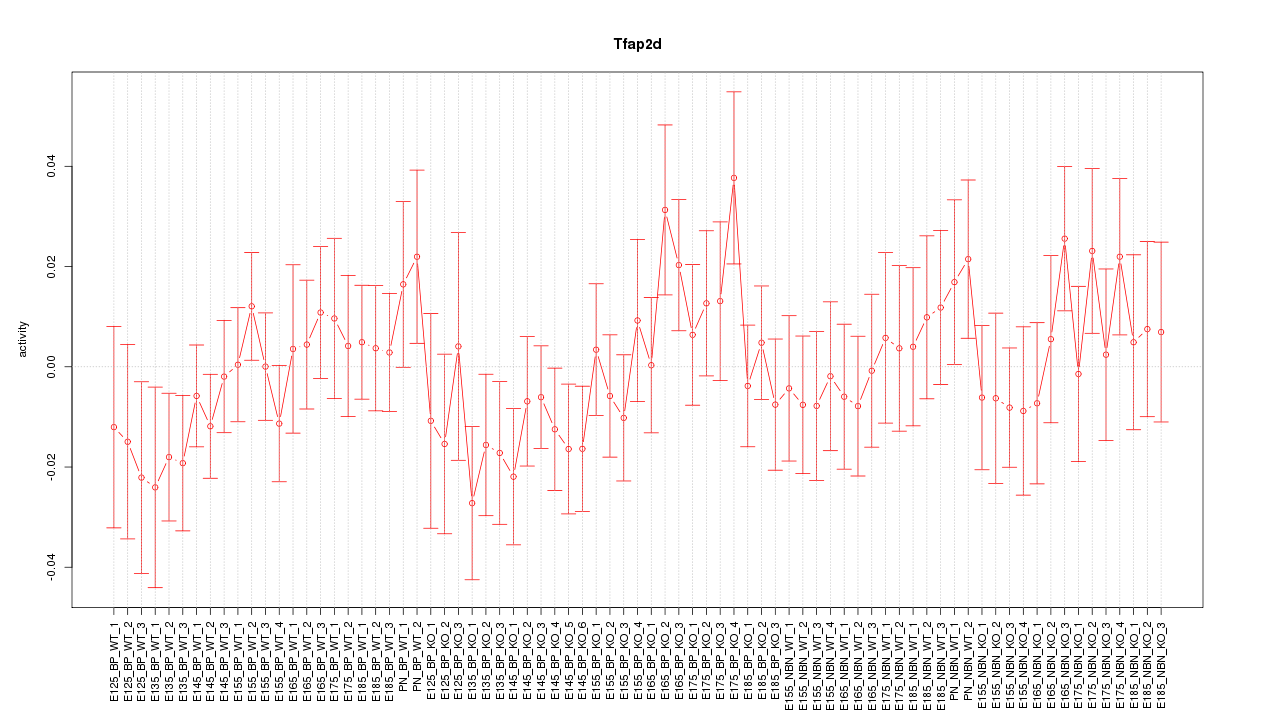
Motif ID: Tfap2d
Z-value: 0.873

Transcription factors associated with Tfap2d:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tfap2d | ENSMUSG00000042596.7 | Tfap2d |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2d | mm10_v2_chr1_+_19103022_19103043 | 0.36 | 1.4e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 1.2 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 1.2 | 7.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 1.1 | 3.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 1.1 | 3.3 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 1.0 | 5.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.9 | 2.8 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.9 | 3.6 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.7 | 3.7 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.7 | 2.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.7 | 2.8 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.6 | 2.5 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.6 | 5.3 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.6 | 1.8 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.6 | 1.7 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.5 | 2.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.5 | 2.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.5 | 4.4 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.5 | 3.9 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.5 | 1.9 | GO:0018307 | tRNA wobble position uridine thiolation(GO:0002143) enzyme active site formation(GO:0018307) |
| 0.5 | 1.9 | GO:0097168 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.5 | 1.4 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.4 | 2.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.4 | 1.6 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.4 | 0.4 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.4 | 1.2 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.4 | 1.2 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.4 | 1.6 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.4 | 1.5 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.4 | 5.7 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.4 | 1.9 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.4 | 3.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.4 | 2.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) |
| 0.4 | 1.1 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.4 | 2.1 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.4 | 2.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) choline metabolic process(GO:0019695) |
| 0.3 | 1.6 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.3 | 1.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 | 0.9 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.3 | 1.5 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.3 | 1.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.3 | 2.7 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.3 | 1.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.3 | 7.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 1.2 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.2 | 1.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 2.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 2.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 3.0 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.2 | 9.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.2 | 0.7 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.2 | 0.7 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.2 | 2.6 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.2 | 2.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 1.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 0.9 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 | 2.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 1.4 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 2.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 | 2.0 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.2 | 9.7 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.2 | 0.8 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 9.4 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 | 1.5 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.7 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.1 | 0.4 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.9 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 4.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 1.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.6 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.4 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.8 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 1.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 2.5 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.1 | 2.1 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 1.0 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 0.6 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 2.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 3.2 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 1.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.5 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 1.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.7 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.3 | GO:1903519 | penetration of zona pellucida(GO:0007341) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.9 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 4.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.4 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 3.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.5 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.9 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.8 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.4 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 1.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.9 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.6 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.1 | 0.7 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 0.8 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 0.3 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.1 | 1.1 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.1 | 1.3 | GO:1901032 | negative regulation of response to reactive oxygen species(GO:1901032) negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.1 | 0.7 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.2 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.8 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.2 | GO:0001787 | natural killer cell proliferation(GO:0001787) NK T cell activation(GO:0051132) |
| 0.1 | 0.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 1.4 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.1 | 0.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.8 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 1.3 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 2.9 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.6 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 1.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.4 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.6 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 2.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 1.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 1.2 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.5 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.8 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 1.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 1.0 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 1.3 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.5 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.9 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 0.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0030934 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 1.0 | 3.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 1.0 | 2.9 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.7 | 3.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.5 | 3.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.4 | 2.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.3 | 1.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 1.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 3.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.3 | 2.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.2 | 3.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 0.9 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 1.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 1.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 1.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 2.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.5 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 3.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.7 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.1 | 1.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 3.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 7.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 2.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 1.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 3.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 1.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.9 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 1.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 2.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 4.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 5.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.4 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 6.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 9.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 5.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 2.9 | GO:0044297 | cell body(GO:0044297) |
| 0.0 | 0.4 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 1.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 2.3 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 2.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 1.0 | 3.1 | GO:0034190 | very-low-density lipoprotein particle binding(GO:0034189) apolipoprotein receptor binding(GO:0034190) |
| 1.0 | 16.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.7 | 2.7 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.6 | 3.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.5 | 2.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.5 | 2.1 | GO:0004104 | choline kinase activity(GO:0004103) cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.5 | 2.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.5 | 3.4 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.4 | 1.9 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.4 | 1.5 | GO:2001070 | starch binding(GO:2001070) |
| 0.4 | 2.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 2.2 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.4 | 1.5 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.3 | 1.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 4.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 1.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 4.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 1.8 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 2.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 2.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.3 | 1.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.2 | 5.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 1.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 3.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 1.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 2.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 0.9 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 1.5 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 1.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 6.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 0.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.2 | 1.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 1.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.2 | 2.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.2 | 5.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 0.7 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.1 | 0.8 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 4.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 2.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 8.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 1.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.1 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 1.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 5.4 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.8 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 1.0 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 2.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.3 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.5 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) mRNA CDS binding(GO:1990715) |
| 0.1 | 0.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.1 | 2.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.8 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 2.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.1 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.9 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 4.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.2 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 2.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.8 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.4 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 2.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 2.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.1 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 5.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.4 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 1.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 1.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.9 | GO:0008144 | drug binding(GO:0008144) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.2 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 5.0 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 6.9 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.1 | 3.5 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.1 | 1.5 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 4.3 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.9 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 2.6 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.7 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 9.6 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.3 | ST_PAC1_RECEPTOR_PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 2.2 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.6 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.3 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.0 | 1.9 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.5 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.4 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.7 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.7 | ST_B_CELL_ANTIGEN_RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 2.6 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.5 | PID_FCER1_PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.7 | ST_FAS_SIGNALING_PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.8 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.5 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.6 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.4 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.0 | 3.5 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID_ERBB1_DOWNSTREAM_PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.5 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 0.9 | REACTOME_DESTABILIZATION_OF_MRNA_BY_BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.3 | 5.0 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.3 | 4.1 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 3.4 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 0.7 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.2 | 2.7 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 3.4 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 12.2 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.2 | 2.1 | REACTOME_SYNTHESIS_OF_PE | Genes involved in Synthesis of PE |
| 0.2 | 3.4 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 7.2 | REACTOME_PI3K_AKT_ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.1 | 2.3 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.1 | 4.7 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 2.4 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 1.2 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 3.4 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.1 | 1.3 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.8 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.5 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.4 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.0 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.5 | REACTOME_PHOSPHORYLATION_OF_CD3_AND_TCR_ZETA_CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.1 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.4 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.2 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 3.5 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.9 | REACTOME_INWARDLY_RECTIFYING_K_CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.4 | REACTOME_ELONGATION_ARREST_AND_RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.1 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 3.0 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.7 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 2.6 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.9 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 3.2 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.0 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 5.1 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.8 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.2 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME_AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |