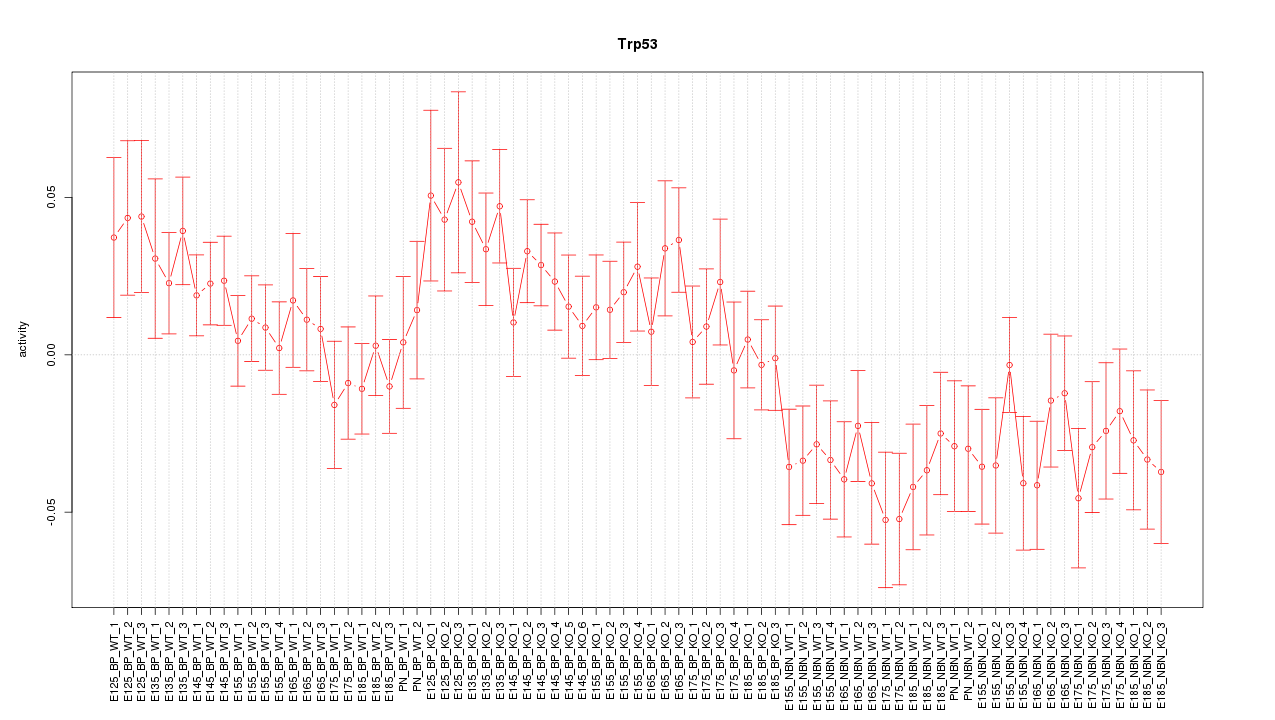
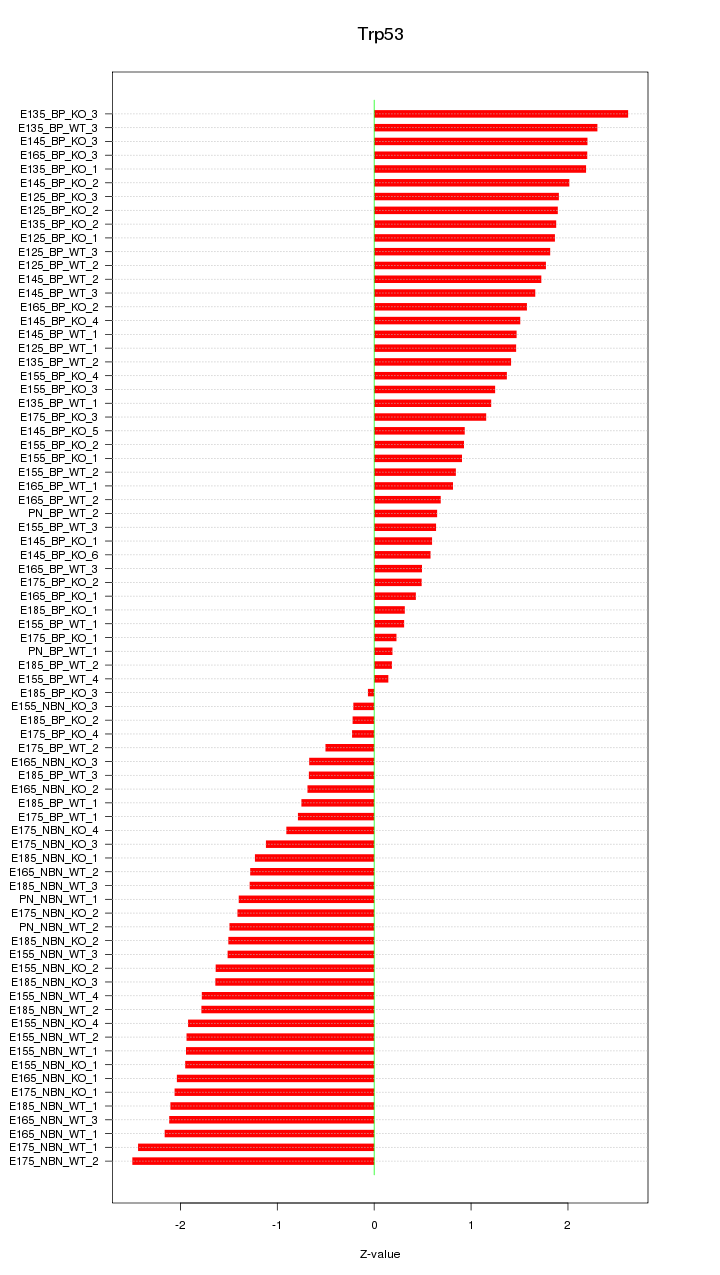
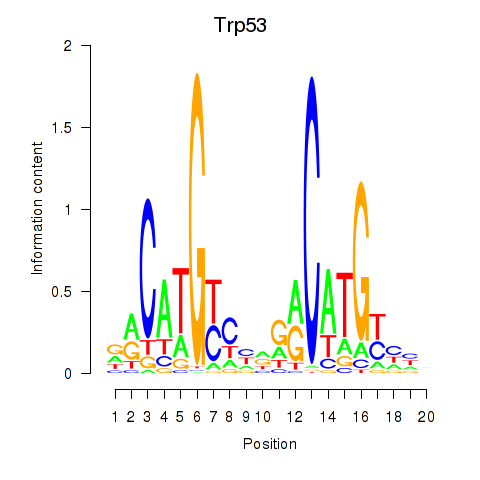
Motif ID: Trp53
Z-value: 1.454
Transcription factors associated with Trp53:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Trp53 | ENSMUSG00000059552.7 | Trp53 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Trp53 | mm10_v2_chr11_+_69580392_69580419 | 0.32 | 5.2e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.4 | 52.2 | GO:0072382 | minus-end-directed vesicle transport along microtubule(GO:0072382) |
| 3.5 | 10.5 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 3.4 | 30.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 2.6 | 7.8 | GO:0002865 | immune response-inhibiting signal transduction(GO:0002765) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 2.4 | 28.8 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 2.1 | 8.6 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 2.1 | 10.6 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 2.1 | 8.3 | GO:0046073 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 1.7 | 18.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 1.6 | 23.8 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 1.3 | 4.0 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 1.3 | 6.6 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 1.2 | 7.3 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 1.2 | 8.2 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 1.0 | 15.7 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.9 | 2.7 | GO:1990046 | positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 0.9 | 6.0 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.7 | 5.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.7 | 16.0 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.7 | 3.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.7 | 2.0 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.7 | 6.0 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.6 | 2.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.6 | 2.5 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.6 | 9.3 | GO:0099623 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) regulation of cardiac muscle cell membrane repolarization(GO:0099623) |
| 0.6 | 61.7 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.5 | 1.6 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.5 | 32.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.4 | 12.9 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.4 | 2.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.4 | 4.6 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.4 | 1.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.4 | 6.2 | GO:0035337 | fatty-acyl-CoA metabolic process(GO:0035337) |
| 0.3 | 1.4 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.3 | 6.0 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.3 | 4.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 1.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 3.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.2 | 1.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 2.0 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 1.2 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.2 | 3.9 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.5 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 2.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 7.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 2.6 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.1 | 4.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 1.9 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 1.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.3 | GO:0035137 | embryonic hindlimb morphogenesis(GO:0035116) hindlimb morphogenesis(GO:0035137) |
| 0.1 | 4.1 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.1 | 2.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.9 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 3.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 0.5 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 1.0 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 4.5 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 8.8 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 1.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.6 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.5 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 3.0 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 2.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 4.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0070102 | interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 4.2 | GO:0042176 | regulation of protein catabolic process(GO:0042176) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.3 | 61.7 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 8.2 | 24.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 6.0 | 23.8 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 3.2 | 16.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 2.7 | 54.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 1.6 | 38.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.5 | 18.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.0 | 6.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.8 | 10.5 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.8 | 8.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.7 | 10.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.7 | 2.0 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.5 | 33.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.4 | 6.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.3 | 4.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.3 | 3.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.3 | 3.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 33.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.2 | 2.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.2 | 10.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 6.6 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 8.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 1.2 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 5.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 8.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 17.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 6.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 3.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 6.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 10.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 27.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 5.6 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.2 | 24.6 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 3.7 | 52.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 3.7 | 18.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 2.3 | 16.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.9 | 5.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.5 | 10.7 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.2 | 3.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 1.0 | 23.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 1.0 | 8.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 1.0 | 6.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.9 | 8.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.8 | 2.5 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.7 | 6.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.7 | 7.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.7 | 4.0 | GO:0050786 | arachidonic acid binding(GO:0050544) RAGE receptor binding(GO:0050786) |
| 0.6 | 2.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.6 | 8.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.5 | 3.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.4 | 4.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.4 | 12.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 1.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.4 | 2.7 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.4 | 24.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 2.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 0.9 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 4.6 | GO:0016594 | glycine binding(GO:0016594) |
| 0.3 | 1.1 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 4.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 4.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 1.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 6.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 0.6 | GO:0019961 | interferon binding(GO:0019961) |
| 0.2 | 2.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.2 | 8.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 34.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 0.5 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 1.3 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.9 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 2.0 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 1.0 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 7.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 7.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 9.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 0.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 2.0 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 5.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 2.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 30.8 | GO:0005215 | transporter activity(GO:0005215) |
| 0.0 | 1.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.6 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 2.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.2 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 4.6 | GO:0003682 | chromatin binding(GO:0003682) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 24.6 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.4 | 8.4 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.4 | 18.4 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 6.7 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 7.5 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.1 | 7.6 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.1 | 7.3 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 9.9 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 6.0 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.4 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.2 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.7 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 52.2 | REACTOME_KINESINS | Genes involved in Kinesins |
| 1.8 | 16.0 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.9 | 24.6 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.6 | 62.7 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.6 | 5.8 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.4 | 8.3 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.4 | 18.4 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 3.5 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.3 | 1.4 | REACTOME_REGULATION_OF_COMPLEMENT_CASCADE | Genes involved in Regulation of Complement cascade |
| 0.3 | 4.1 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 23.8 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 10.6 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 4.6 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.2 | 4.5 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.0 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 7.3 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.2 | REACTOME_NEP_NS2_INTERACTS_WITH_THE_CELLULAR_EXPORT_MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 3.2 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.7 | REACTOME_CROSS_PRESENTATION_OF_SOLUBLE_EXOGENOUS_ANTIGENS_ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.1 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.4 | REACTOME_PACKAGING_OF_TELOMERE_ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.6 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.9 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 6.6 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.3 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.0 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.1 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |