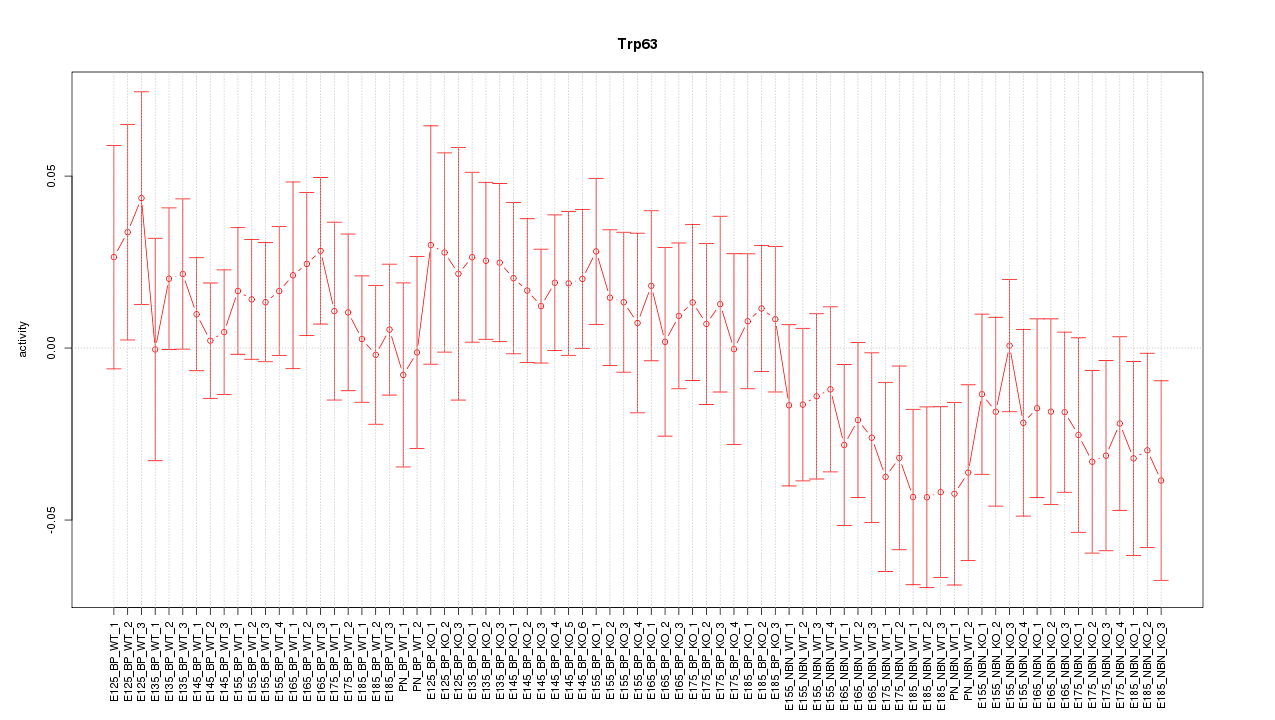
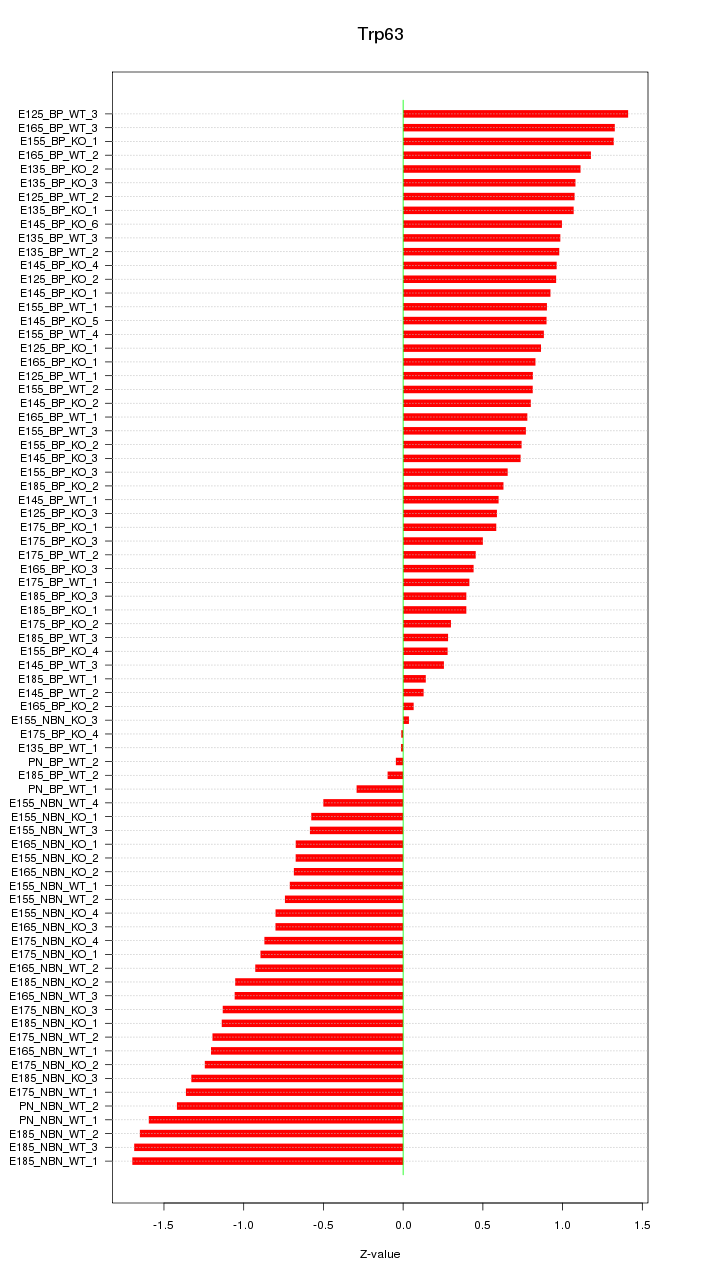
Motif ID: Trp63
Z-value: 0.895

Transcription factors associated with Trp63:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Trp63 | ENSMUSG00000022510.8 | Trp63 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 15.6 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 2.3 | 11.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 2.0 | 10.0 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 1.8 | 20.0 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 1.6 | 14.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.9 | 3.4 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.7 | 4.9 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.4 | 6.9 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 23.5 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.3 | 1.0 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.3 | 3.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.3 | 6.9 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.3 | 2.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.3 | 3.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 6.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 2.7 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.2 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 3.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.6 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 1.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 9.2 | GO:0000278 | mitotic cell cycle(GO:0000278) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 2.6 | 23.5 | GO:0030478 | actin cap(GO:0030478) |
| 2.0 | 10.0 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.5 | 3.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.4 | 6.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 4.9 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.5 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 11.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 20.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 2.7 | GO:0043679 | axon terminus(GO:0043679) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.7 | 6.9 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 1.3 | 10.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.5 | 10.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.5 | 6.9 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 3.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 4.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 3.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.2 | 2.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 1.0 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.6 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.1 | 20.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 1.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 3.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 24.2 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 15.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 11.4 | GO:0016491 | oxidoreductase activity(GO:0016491) |
| 0.0 | 10.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 14.2 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 4.9 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.2 | 6.9 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 10.3 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 6.5 | ST_ERK1_ERK2_MAPK_PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.9 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.9 | REACTOME_FACILITATIVE_NA_INDEPENDENT_GLUCOSE_TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.6 | 23.5 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.6 | 3.9 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.5 | 4.9 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.5 | 14.2 | REACTOME_AKT_PHOSPHORYLATES_TARGETS_IN_THE_CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 10.0 | REACTOME_UNFOLDED_PROTEIN_RESPONSE | Genes involved in Unfolded Protein Response |
| 0.1 | 3.2 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.8 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |