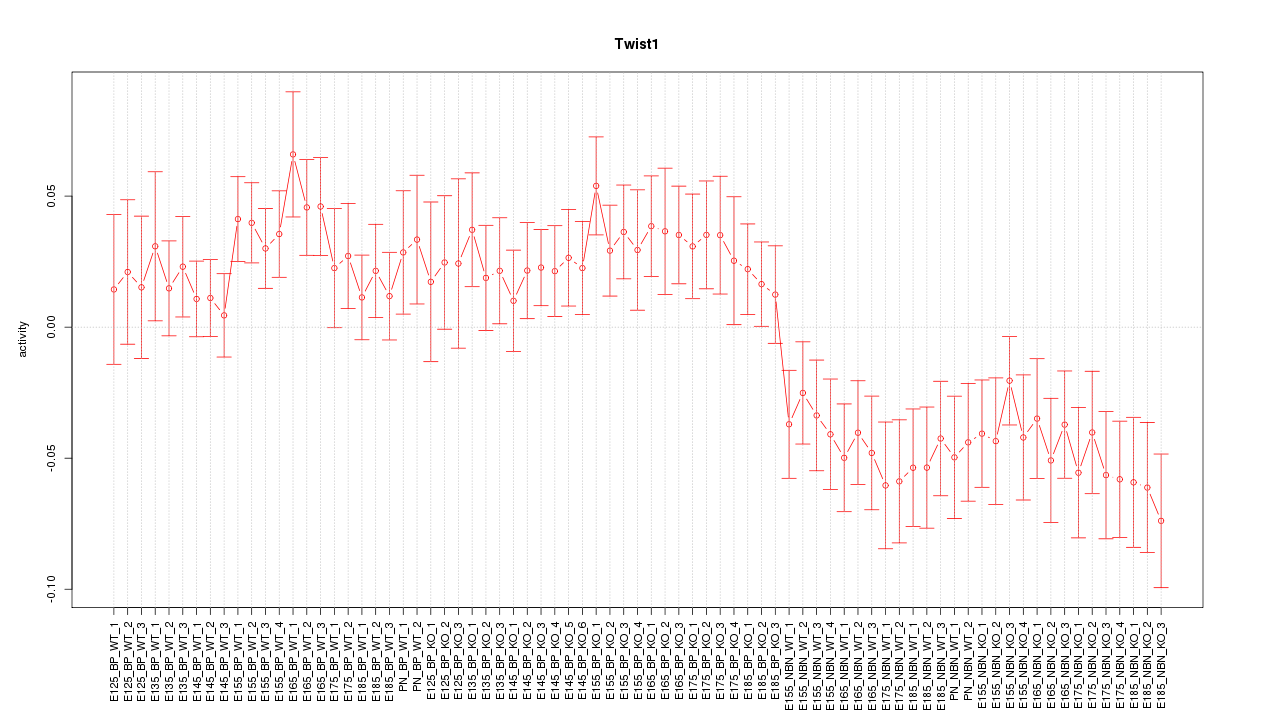
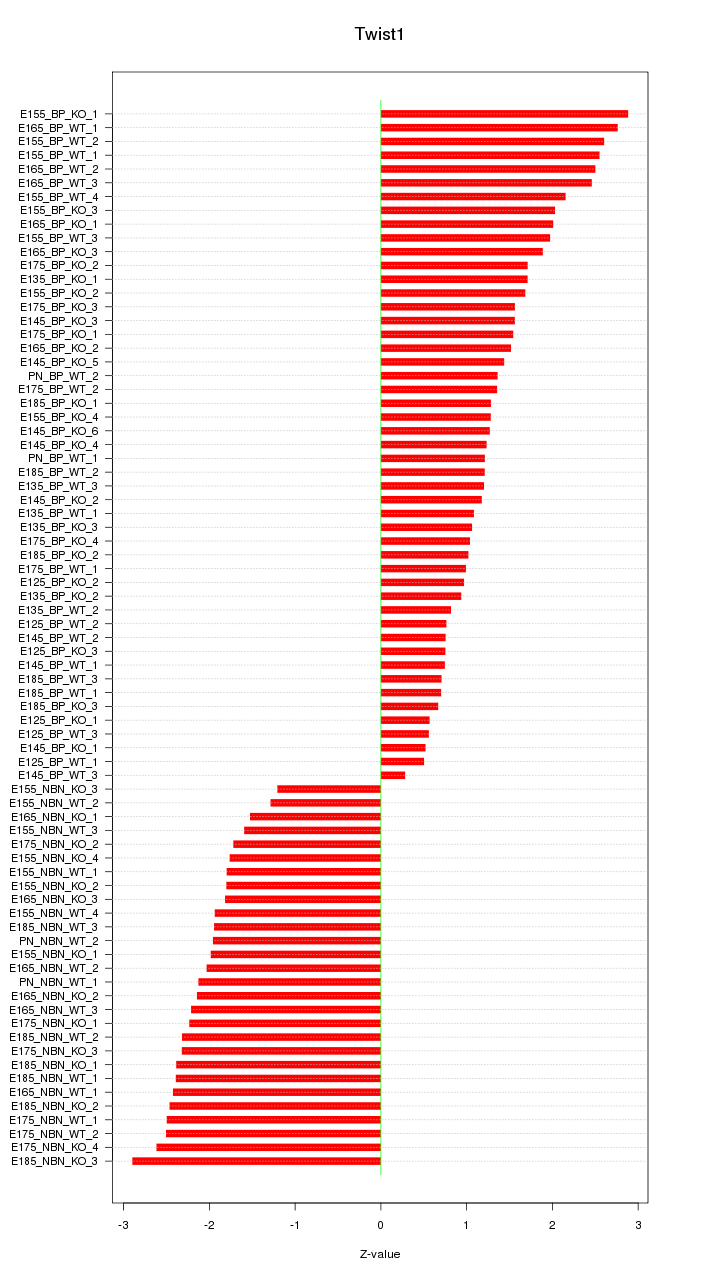
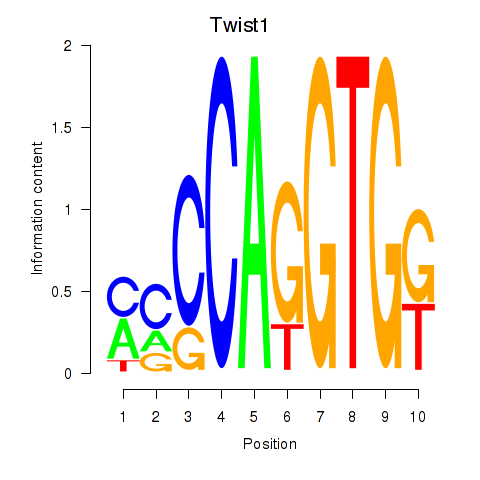
Motif ID: Twist1
Z-value: 1.747
Transcription factors associated with Twist1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Twist1 | ENSMUSG00000035799.5 | Twist1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Twist1 | mm10_v2_chr12_+_33957645_33957671 | -0.20 | 7.9e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 12.6 | 37.7 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 12.5 | 50.0 | GO:0060032 | notochord regression(GO:0060032) |
| 9.6 | 9.6 | GO:0090194 | negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) |
| 7.9 | 31.6 | GO:0051311 | spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 6.9 | 20.7 | GO:1902460 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 4.1 | 37.0 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 3.6 | 3.6 | GO:1904173 | regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 3.6 | 10.8 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 3.1 | 21.7 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 2.6 | 7.8 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 2.5 | 10.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 2.2 | 2.2 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 2.0 | 10.0 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 2.0 | 11.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 1.8 | 16.6 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 1.7 | 5.0 | GO:0061324 | trachea cartilage morphogenesis(GO:0060535) mammary placode formation(GO:0060596) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 1.6 | 4.7 | GO:0030070 | insulin processing(GO:0030070) |
| 1.4 | 9.9 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 1.3 | 24.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 1.3 | 3.9 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 1.3 | 6.4 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 1.2 | 15.9 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 1.2 | 3.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 1.2 | 15.0 | GO:0001675 | acrosome assembly(GO:0001675) |
| 1.1 | 5.6 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 1.1 | 38.5 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 1.0 | 19.9 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 1.0 | 7.1 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.9 | 10.0 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.8 | 2.5 | GO:2000508 | regulation of dendritic cell chemotaxis(GO:2000508) |
| 0.8 | 6.9 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.7 | 8.8 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.6 | 2.8 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.5 | 31.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.5 | 1.5 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.5 | 1.4 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.4 | 1.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 5.8 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.3 | 1.3 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.3 | 10.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.3 | 5.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 1.6 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.2 | 1.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.2 | 2.4 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 | 1.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.2 | 2.4 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.2 | 1.9 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 2.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 2.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 4.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 4.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.4 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 4.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 2.9 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.1 | 2.0 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.6 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 8.8 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 8.7 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.5 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 5.0 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 6.9 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.4 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.8 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 1.5 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 2.4 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 5.2 | GO:0010975 | regulation of neuron projection development(GO:0010975) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.3 | 33.3 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 6.6 | 19.9 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 6.1 | 24.2 | GO:0043293 | apoptosome(GO:0043293) |
| 3.8 | 50.0 | GO:0097542 | ciliary tip(GO:0097542) |
| 3.6 | 10.8 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 3.5 | 31.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 2.8 | 16.6 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 2.4 | 7.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 1.3 | 5.0 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.9 | 2.6 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.8 | 15.0 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.7 | 5.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.7 | 5.8 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.6 | 1.8 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.6 | 7.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.4 | 6.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.3 | 4.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 2.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 2.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 4.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 3.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 39.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.6 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 2.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 9.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 8.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 1.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 3.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 5.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 10.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 4.7 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 2.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 5.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.5 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 19.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 82.5 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.2 | GO:0031902 | late endosome membrane(GO:0031902) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.6 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 4.2 | 12.7 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 1.7 | 9.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 1.5 | 21.7 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 1.5 | 8.8 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.4 | 10.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 1.3 | 3.9 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 1.2 | 42.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 1.1 | 19.9 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 1.0 | 9.6 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.9 | 6.0 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.9 | 6.9 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.8 | 20.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.8 | 10.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.8 | 10.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.8 | 10.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.7 | 2.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.7 | 5.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 37.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.5 | 17.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.5 | 4.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 7.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 10.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 5.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.4 | 1.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.4 | 3.6 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 3.5 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.3 | 10.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.3 | 2.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.3 | 7.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.3 | 33.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.3 | 5.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 7.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 9.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 10.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.2 | 6.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 2.6 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.2 | 0.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 1.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 4.5 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 2.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 31.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.1 | 2.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 1.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.4 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.1 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 6.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 23.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 1.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 5.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 3.0 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 33.3 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.5 | 48.8 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.9 | 9.6 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.7 | 31.6 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.5 | 33.9 | PID_CD8_TCR_DOWNSTREAM_PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.5 | 14.9 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.3 | 10.9 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.2 | 22.2 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.2 | 10.0 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.2 | 29.7 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.3 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 2.4 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 5.0 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 6.4 | PID_TAP63_PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.7 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.1 | 3.9 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.1 | 2.6 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.9 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | PID_INTEGRIN1_PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.2 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.0 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 9.9 | REACTOME_SYNTHESIS_OF_BILE_ACIDS_AND_BILE_SALTS_VIA_24_HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.8 | 16.6 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.7 | 10.8 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.6 | 19.9 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.6 | 6.4 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.5 | 10.0 | REACTOME_DESTABILIZATION_OF_MRNA_BY_TRISTETRAPROLIN_TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.5 | 6.6 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.4 | 8.8 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.4 | 3.9 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.4 | 31.9 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.3 | 7.5 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 19.3 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 4.2 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.2 | 2.3 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 3.0 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 4.7 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.2 | 1.1 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.3 | REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 5.0 | REACTOME_NUCLEAR_EVENTS_KINASE_AND_TRANSCRIPTION_FACTOR_ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.1 | 10.0 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 3.6 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 3.5 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.4 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 10.2 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 2.8 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 2.1 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.5 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |