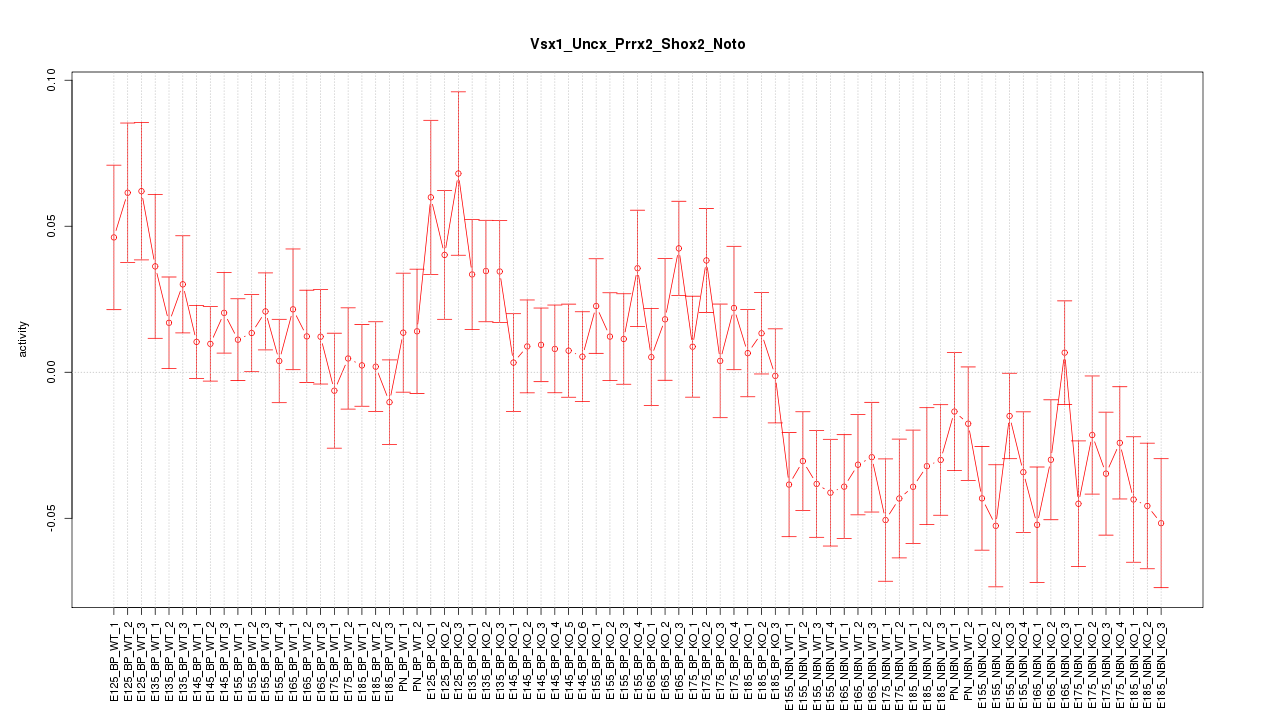
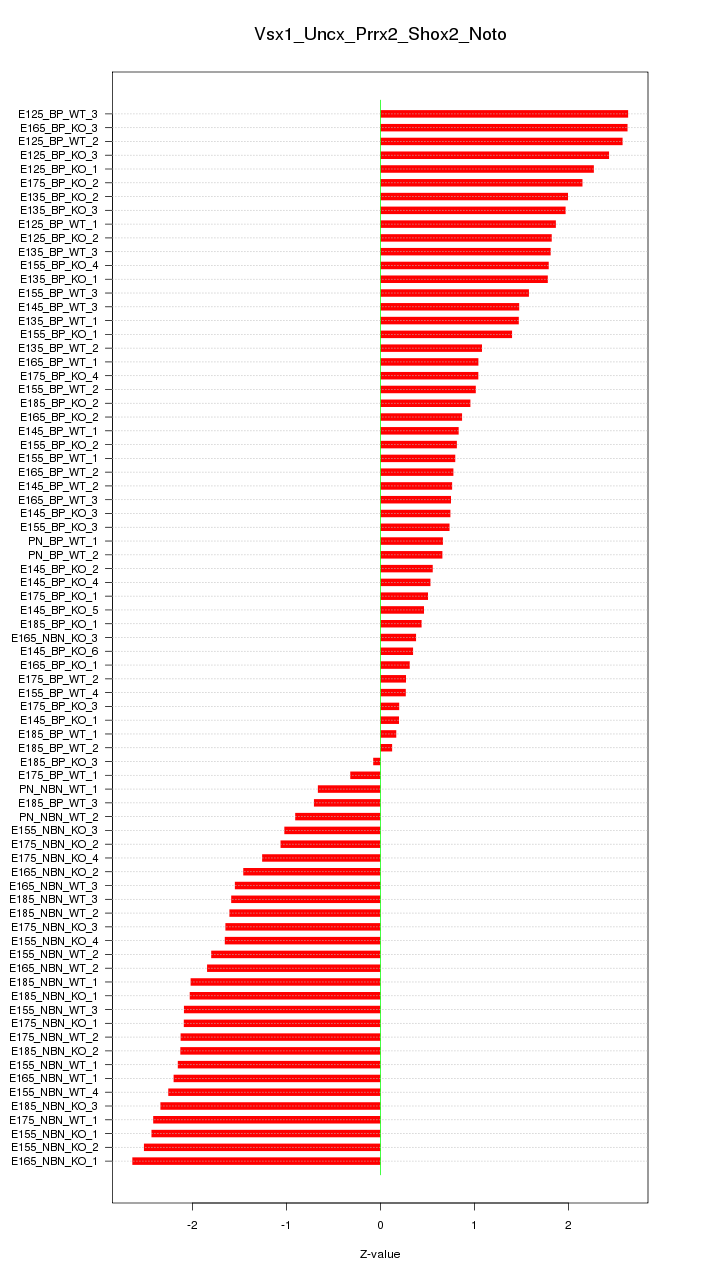
Motif ID: Vsx1_Uncx_Prrx2_Shox2_Noto
Z-value: 1.535
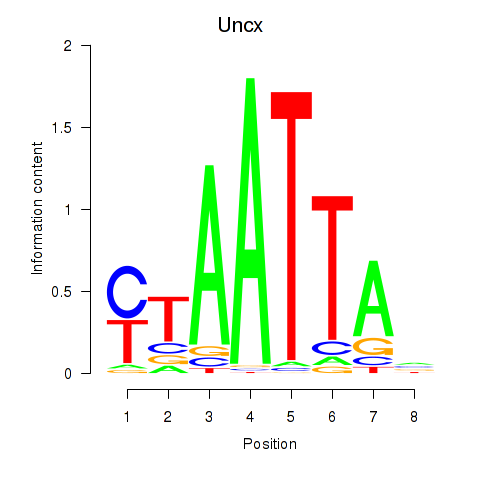
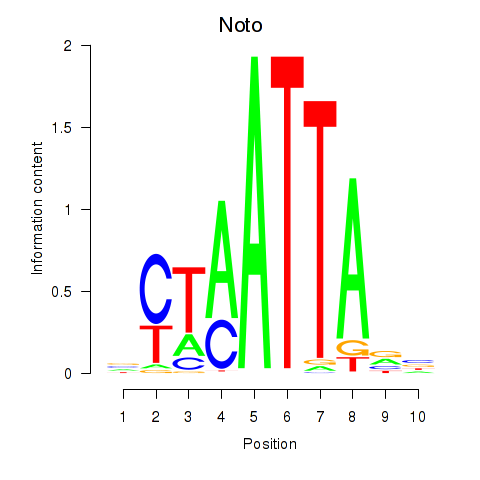
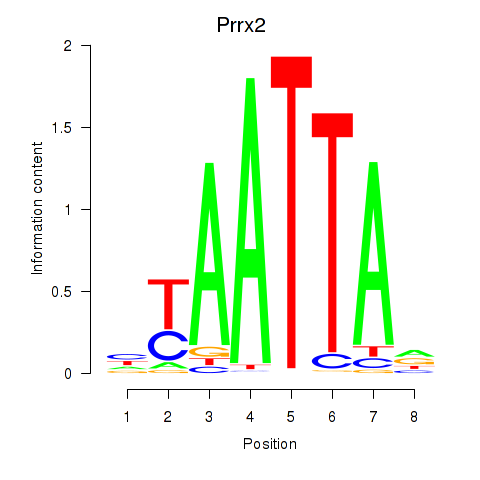
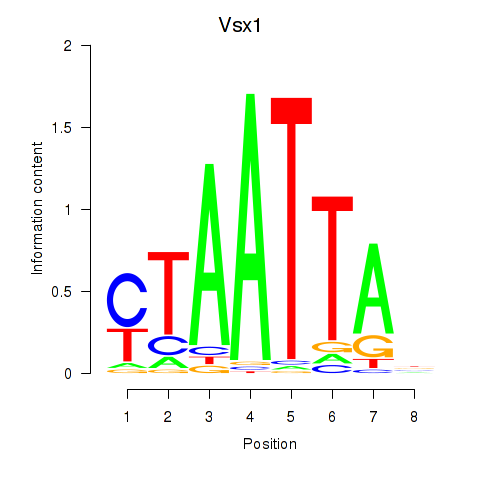
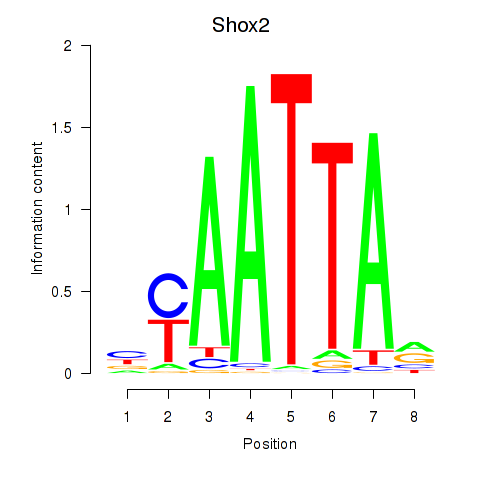
Transcription factors associated with Vsx1_Uncx_Prrx2_Shox2_Noto:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Noto | ENSMUSG00000068302.7 | Noto |
| Prrx2 | ENSMUSG00000039476.7 | Prrx2 |
| Shox2 | ENSMUSG00000027833.10 | Shox2 |
| Uncx | ENSMUSG00000029546.11 | Uncx |
| Vsx1 | ENSMUSG00000033080.8 | Vsx1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Uncx | mm10_v2_chr5_+_139543889_139543904 | 0.75 | 7.8e-15 | Click! |
| Shox2 | mm10_v2_chr3_-_66981279_66981318 | 0.32 | 4.0e-03 | Click! |
| Prrx2 | mm10_v2_chr2_+_30845059_30845059 | -0.07 | 5.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 57.0 | 170.9 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 10.2 | 30.7 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 10.1 | 80.7 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 9.1 | 100.6 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 8.8 | 35.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 8.2 | 24.6 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 6.4 | 19.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 5.4 | 5.4 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 4.2 | 12.7 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 4.1 | 24.8 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 3.9 | 19.4 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 3.5 | 14.1 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 3.4 | 54.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 3.2 | 9.7 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 2.7 | 50.6 | GO:0071803 | keratinocyte development(GO:0003334) positive regulation of podosome assembly(GO:0071803) |
| 2.6 | 18.4 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 2.5 | 7.6 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 2.4 | 31.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 2.4 | 11.8 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 2.2 | 28.9 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 2.1 | 4.3 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) |
| 2.1 | 6.4 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 2.1 | 8.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 2.0 | 4.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 2.0 | 35.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 1.8 | 16.2 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 1.7 | 15.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 1.7 | 5.0 | GO:0036233 | glycine import(GO:0036233) |
| 1.7 | 5.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 1.6 | 8.0 | GO:0001661 | conditioned taste aversion(GO:0001661) amygdala development(GO:0021764) |
| 1.6 | 14.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 1.6 | 6.3 | GO:0060729 | pancreatic A cell differentiation(GO:0003310) intestinal epithelial structure maintenance(GO:0060729) |
| 1.5 | 18.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.5 | 4.5 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 1.5 | 4.4 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 1.4 | 12.8 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 1.4 | 19.5 | GO:0060013 | righting reflex(GO:0060013) |
| 1.4 | 63.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 1.3 | 14.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 1.2 | 3.6 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.2 | 3.6 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 1.2 | 14.2 | GO:0072189 | ureter development(GO:0072189) |
| 1.1 | 21.6 | GO:0007530 | sex determination(GO:0007530) |
| 1.0 | 24.6 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 1.0 | 7.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 1.0 | 2.9 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.9 | 7.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.9 | 1.7 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.9 | 4.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.8 | 4.0 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation by host of viral process(GO:0044793) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.7 | 10.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.7 | 1.4 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.7 | 15.0 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of mitotic spindle localization(GO:0040001) |
| 0.7 | 5.6 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.6 | 1.9 | GO:0050748 | negative regulation of receptor biosynthetic process(GO:0010871) transforming growth factor-beta secretion(GO:0038044) negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.6 | 2.5 | GO:0061743 | motor learning(GO:0061743) |
| 0.6 | 9.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.6 | 1.8 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.6 | 3.5 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.6 | 4.6 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.6 | 4.4 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.6 | 0.6 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.5 | 10.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.4 | 3.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.4 | 1.2 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 0.4 | 2.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.4 | 1.9 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.4 | 1.8 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.4 | 1.1 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.4 | 13.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.4 | 1.8 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.4 | 1.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.4 | 17.6 | GO:0030901 | midbrain development(GO:0030901) |
| 0.3 | 2.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 1.4 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 1.7 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.3 | 1.9 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.3 | 0.9 | GO:0060023 | soft palate development(GO:0060023) |
| 0.3 | 1.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 1.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.3 | 2.8 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.3 | 1.4 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.3 | 1.9 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) |
| 0.3 | 1.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.3 | 6.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 9.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 0.3 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.2 | 2.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 7.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.2 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.2 | 2.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.2 | 3.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 0.5 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.2 | 1.4 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.2 | 2.8 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 25.0 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.2 | 12.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.2 | 0.9 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.2 | 3.2 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) |
| 0.2 | 7.1 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.2 | 5.2 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 2.0 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.2 | 18.5 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.2 | 0.8 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 0.9 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 1.2 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.2 | 2.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.2 | 0.7 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.2 | 4.2 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.2 | 0.8 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 | 1.6 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 7.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.2 | 0.8 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 | 2.0 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 1.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 24.5 | GO:0022409 | positive regulation of cell-cell adhesion(GO:0022409) |
| 0.1 | 0.9 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 4.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 2.5 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.4 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 1.2 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.1 | 4.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.4 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.1 | 1.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 4.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.6 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.1 | 0.4 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.9 | GO:0061013 | regulation of mRNA catabolic process(GO:0061013) positive regulation of mRNA catabolic process(GO:0061014) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 2.8 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 | 0.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.6 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 2.5 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 3.9 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 2.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 3.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 1.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 13.4 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 1.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 5.3 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 8.6 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 1.9 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) |
| 0.0 | 2.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.7 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 15.5 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 3.2 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.2 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 1.0 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.9 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 1.4 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 24.6 | GO:0060187 | cell pole(GO:0060187) |
| 3.1 | 18.6 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 1.5 | 9.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 1.3 | 6.4 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 1.2 | 18.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.1 | 50.6 | GO:0002102 | podosome(GO:0002102) |
| 1.0 | 5.0 | GO:0031523 | Myb complex(GO:0031523) |
| 1.0 | 2.9 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.9 | 24.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.8 | 5.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 1.9 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.5 | 3.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.5 | 2.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.5 | 6.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.5 | 3.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.5 | 2.3 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.4 | 24.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.4 | 2.8 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.4 | 3.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.4 | 1.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 1.4 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.4 | 12.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 11.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.3 | 0.9 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 10.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 14.8 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.3 | 5.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 9.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 35.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.2 | 2.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.2 | 4.8 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.2 | 0.9 | GO:0000178 | cytoplasmic exosome (RNase complex)(GO:0000177) exosome (RNase complex)(GO:0000178) |
| 0.2 | 6.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.2 | 8.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 3.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.2 | 1.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 8.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.2 | 70.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.2 | 16.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.2 | 0.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.9 | GO:0051286 | cell tip(GO:0051286) |
| 0.1 | 1.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 2.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 1.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 629.9 | GO:0005634 | nucleus(GO:0005634) |
| 0.1 | 11.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.1 | 15.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 2.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 14.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 9.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 32.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 4.5 | 67.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 3.8 | 30.7 | GO:0050693 | LBD domain binding(GO:0050693) |
| 3.5 | 14.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 3.0 | 9.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 2.7 | 177.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 2.0 | 18.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 2.0 | 15.9 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 1.9 | 19.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 1.9 | 24.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 1.8 | 3.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.6 | 15.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 1.6 | 18.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.5 | 4.6 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 1.4 | 4.2 | GO:0005118 | sevenless binding(GO:0005118) |
| 1.2 | 8.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 1.2 | 13.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 1.1 | 7.7 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.1 | 7.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.0 | 4.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.9 | 3.6 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.8 | 19.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.7 | 21.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.6 | 1.9 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.6 | 20.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.6 | 3.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.6 | 5.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.6 | 194.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.5 | 10.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.5 | 18.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.5 | 5.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.5 | 23.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.5 | 3.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.4 | 3.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.4 | 2.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.4 | 2.8 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.4 | 7.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 1.9 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) neuregulin binding(GO:0038132) |
| 0.4 | 8.1 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 10.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 1.4 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.4 | 2.9 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.3 | 3.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 9.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.3 | 42.4 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.3 | 273.0 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.3 | 17.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.3 | 0.9 | GO:1904047 | nitrite reductase activity(GO:0098809) S-adenosyl-L-methionine binding(GO:1904047) |
| 0.3 | 16.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 0.6 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.3 | 1.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.2 | 6.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 1.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.9 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 0.9 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.2 | 1.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.2 | 4.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.2 | 9.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 3.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.2 | 1.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 4.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 1.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 1.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 0.8 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 1.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 2.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 3.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.4 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.9 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.1 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 7.1 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.1 | 0.8 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.1 | 3.3 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 1.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 13.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 4.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 2.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 17.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 1.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 2.7 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 1.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 2.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 36.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 1.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 4.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.3 | GO:0047485 | protein N-terminus binding(GO:0047485) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 104.1 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 2.9 | 170.9 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.7 | 15.9 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.7 | 21.6 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.7 | 32.0 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 0.7 | 19.3 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.6 | 23.3 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.6 | 24.0 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.3 | 30.9 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.3 | 14.4 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.3 | 9.6 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.3 | 12.6 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 3.7 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.0 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 4.0 | ST_ADRENERGIC | Adrenergic Pathway |
| 0.2 | 2.2 | PID_SYNDECAN_3_PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.1 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 2.2 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 9.1 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.1 | 10.8 | PID_MYC_ACTIV_PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.0 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.1 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 6.1 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 2.8 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.0 | 2.4 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.0 | 1.9 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 2.2 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.4 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.2 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.2 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.3 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 32.3 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.7 | 6.3 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.5 | 5.1 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 37.4 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.4 | 2.2 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 4.6 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 3.7 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.4 | 24.2 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.4 | 6.4 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.3 | 7.6 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 12.6 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.2 | 1.9 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.2 | 20.2 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 12.7 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 31.0 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 4.4 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 6.1 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 2.7 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 2.2 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 2.8 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 4.8 | REACTOME_RNA_POL_II_TRANSCRIPTION_PRE_INITIATION_AND_PROMOTER_OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.2 | 2.9 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 1.3 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 6.7 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 4.1 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.1 | 3.7 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.8 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.1 | 2.0 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 8.3 | REACTOME_G_ALPHA_Q_SIGNALLING_EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 2.0 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 2.2 | REACTOME_GASTRIN_CREB_SIGNALLING_PATHWAY_VIA_PKC_AND_MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.1 | 0.6 | REACTOME_PROLONGED_ERK_ACTIVATION_EVENTS | Genes involved in Prolonged ERK activation events |
| 0.1 | 1.6 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.7 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.1 | 1.1 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.4 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.1 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.1 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.2 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.9 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME_NOREPINEPHRINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.9 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.0 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |