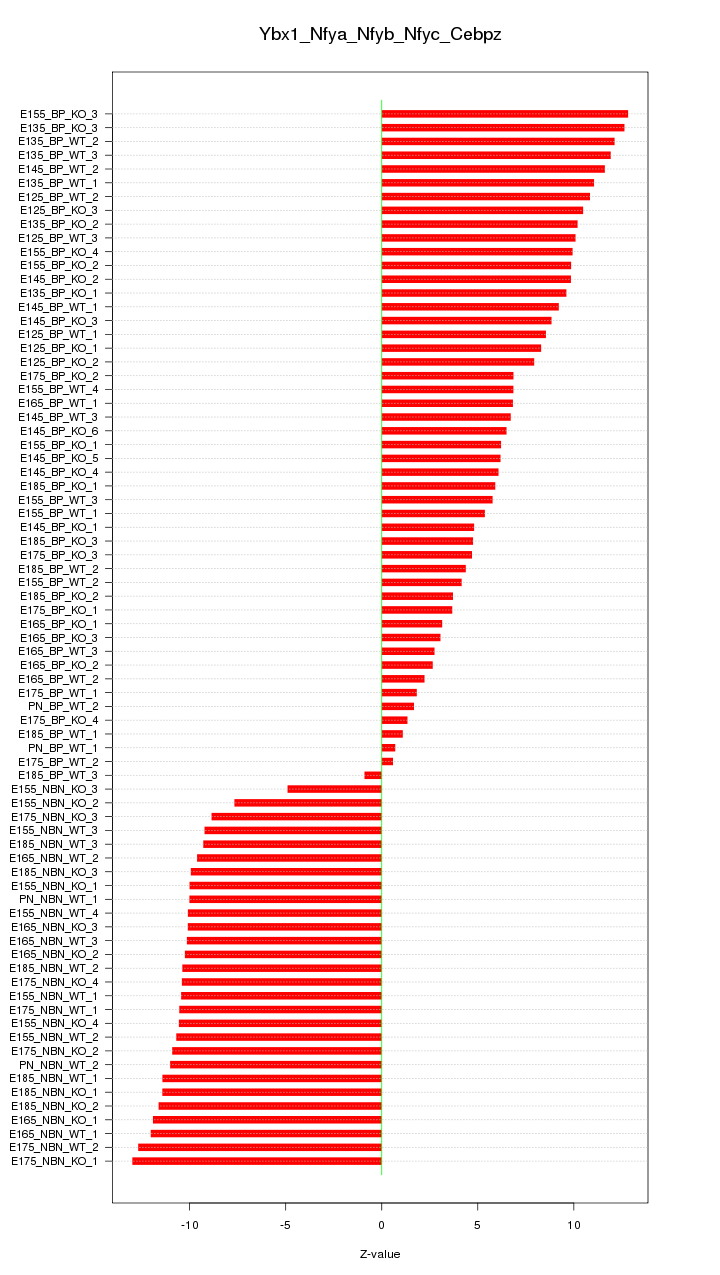
Motif ID: Ybx1_Nfya_Nfyb_Nfyc_Cebpz
Z-value: 8.624

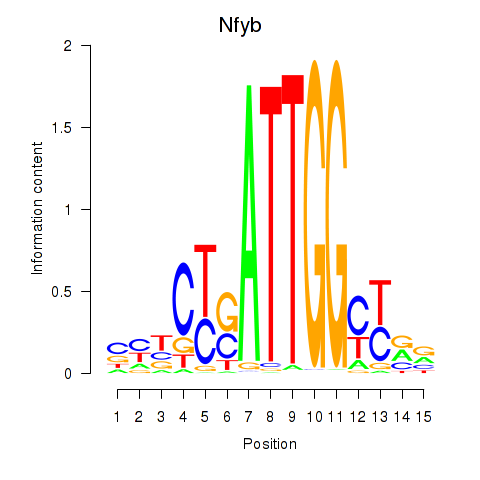
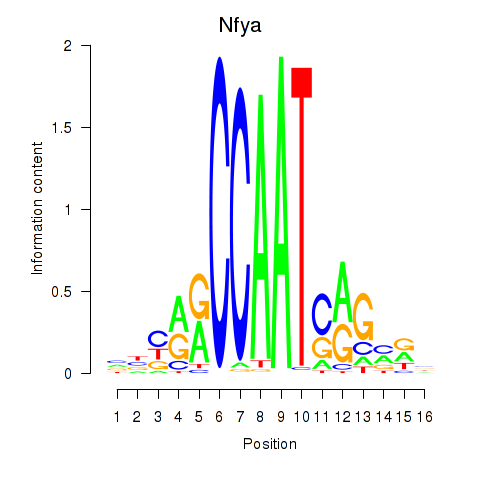
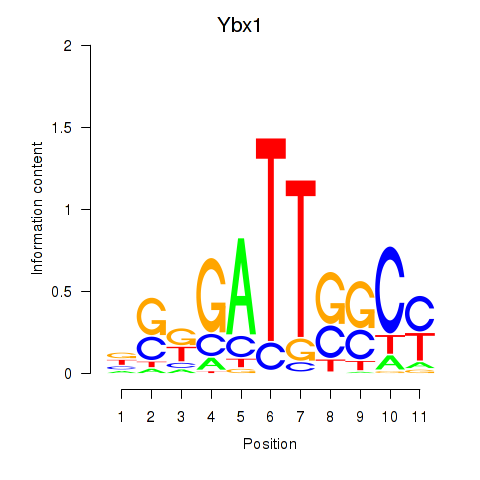
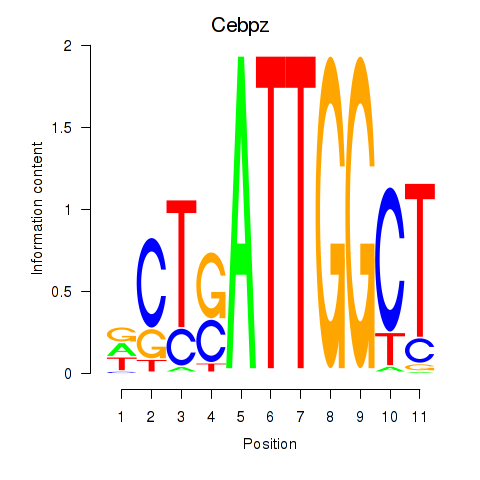
Transcription factors associated with Ybx1_Nfya_Nfyb_Nfyc_Cebpz:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Cebpz | ENSMUSG00000024081.8 | Cebpz |
| Nfya | ENSMUSG00000023994.7 | Nfya |
| Nfyb | ENSMUSG00000020248.12 | Nfyb |
| Nfyc | ENSMUSG00000032897.11 | Nfyc |
| Ybx1 | ENSMUSG00000028639.8 | Ybx1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfyc | mm10_v2_chr4_-_120815703_120815761 | 0.50 | 3.0e-06 | Click! |
| Ybx1 | mm10_v2_chr4_-_119294520_119294604 | -0.35 | 1.6e-03 | Click! |
| Cebpz | mm10_v2_chr17_-_78937031_78937074 | -0.26 | 2.3e-02 | Click! |
| Nfyb | mm10_v2_chr10_-_82764088_82764144 | -0.10 | 3.8e-01 | Click! |
| Nfya | mm10_v2_chr17_-_48409729_48409906 | -0.04 | 7.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 73.3 | 220.0 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 45.5 | 181.8 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 43.3 | 129.9 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 33.1 | 198.7 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 29.4 | 529.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 28.0 | 83.9 | GO:0072382 | minus-end-directed vesicle transport along microtubule(GO:0072382) |
| 27.9 | 55.8 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 26.8 | 80.5 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 25.0 | 74.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 24.0 | 119.8 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 23.4 | 328.2 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 21.2 | 84.9 | GO:1902724 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 21.1 | 84.4 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 20.0 | 139.8 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 19.5 | 58.5 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 18.6 | 111.6 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 17.5 | 17.5 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 17.3 | 52.0 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 16.3 | 49.0 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 16.3 | 32.6 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 15.8 | 47.4 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 15.8 | 47.4 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 15.6 | 140.8 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 15.3 | 30.7 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 15.2 | 45.5 | GO:1990705 | cholangiocyte proliferation(GO:1990705) |
| 14.6 | 58.4 | GO:0021648 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) cranial nerve formation(GO:0021603) vestibulocochlear nerve morphogenesis(GO:0021648) |
| 14.1 | 28.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 13.7 | 123.7 | GO:0033504 | floor plate development(GO:0033504) |
| 13.7 | 41.0 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 13.4 | 214.3 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 13.2 | 52.9 | GO:1903463 | regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 12.9 | 180.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 12.4 | 49.7 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 12.2 | 12.2 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 12.0 | 36.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 12.0 | 35.9 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 11.7 | 82.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 11.6 | 34.8 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 11.4 | 56.9 | GO:2000427 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) positive regulation of apoptotic cell clearance(GO:2000427) |
| 11.3 | 79.4 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 10.1 | 10.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 10.0 | 30.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 10.0 | 311.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 10.0 | 50.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 10.0 | 29.9 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 9.8 | 19.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 9.8 | 29.3 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 9.7 | 48.5 | GO:0060022 | hard palate development(GO:0060022) |
| 9.6 | 95.6 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 9.4 | 28.2 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 8.8 | 26.4 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 8.6 | 112.0 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 8.6 | 25.7 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 8.5 | 51.2 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 8.5 | 67.7 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 8.2 | 24.5 | GO:0046671 | negative regulation of retinal cell programmed cell death(GO:0046671) |
| 7.9 | 15.9 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 7.7 | 46.3 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 7.5 | 217.6 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 7.4 | 36.8 | GO:1990839 | response to endothelin(GO:1990839) |
| 7.3 | 7.3 | GO:0060364 | frontal suture morphogenesis(GO:0060364) |
| 7.3 | 102.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 7.3 | 123.5 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 7.2 | 172.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 7.1 | 78.4 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 7.0 | 28.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 6.9 | 62.3 | GO:0042095 | interferon-gamma biosynthetic process(GO:0042095) |
| 6.9 | 27.4 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 6.8 | 20.5 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 6.8 | 20.5 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 6.8 | 27.0 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 6.7 | 20.2 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 6.7 | 13.5 | GO:0002645 | positive regulation of tolerance induction(GO:0002645) |
| 6.7 | 13.4 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 6.5 | 52.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 6.5 | 6.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 6.5 | 12.9 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 6.2 | 18.6 | GO:1902605 | heterotrimeric G-protein complex assembly(GO:1902605) |
| 6.1 | 18.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 6.1 | 54.7 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 6.0 | 24.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 6.0 | 23.8 | GO:0034239 | regulation of macrophage fusion(GO:0034239) |
| 5.9 | 17.7 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 5.8 | 11.6 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 5.8 | 51.9 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 5.7 | 17.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 5.7 | 17.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 5.6 | 33.7 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 5.6 | 27.8 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 5.4 | 48.8 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 5.4 | 32.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 5.3 | 10.7 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 5.3 | 15.8 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 5.3 | 31.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 5.1 | 5.1 | GO:0016445 | somatic diversification of immunoglobulins(GO:0016445) |
| 5.1 | 20.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 5.0 | 24.9 | GO:0048478 | replication fork protection(GO:0048478) |
| 5.0 | 59.5 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 4.9 | 24.7 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 4.9 | 44.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 4.9 | 73.7 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 4.7 | 18.9 | GO:0006272 | leading strand elongation(GO:0006272) |
| 4.6 | 9.2 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 4.6 | 32.1 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 4.5 | 13.6 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 4.5 | 22.4 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 4.4 | 31.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 4.4 | 39.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 4.4 | 13.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 4.3 | 12.9 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 4.2 | 29.7 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 4.2 | 8.4 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 4.2 | 21.0 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 4.2 | 12.5 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 4.1 | 12.4 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 4.1 | 45.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 4.1 | 8.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 4.1 | 20.5 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 4.1 | 12.2 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 4.0 | 28.3 | GO:0006551 | leucine metabolic process(GO:0006551) |
| 4.0 | 44.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 4.0 | 56.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 4.0 | 16.1 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 4.0 | 64.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 4.0 | 12.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 4.0 | 15.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 3.9 | 7.8 | GO:0051026 | chiasma assembly(GO:0051026) |
| 3.9 | 15.7 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 3.9 | 54.6 | GO:0043486 | histone exchange(GO:0043486) |
| 3.9 | 42.7 | GO:0060539 | diaphragm development(GO:0060539) |
| 3.9 | 15.4 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 3.9 | 3.9 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 3.8 | 95.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 3.8 | 11.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 3.8 | 3.8 | GO:0051299 | centrosome separation(GO:0051299) |
| 3.8 | 15.2 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 3.7 | 22.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 3.7 | 11.0 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 3.6 | 3.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 3.6 | 21.5 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 3.5 | 10.6 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 3.5 | 42.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 3.5 | 7.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 3.5 | 63.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 3.5 | 14.0 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 3.5 | 10.4 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 3.5 | 24.4 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 3.5 | 45.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 3.4 | 27.4 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 3.4 | 65.1 | GO:0007099 | centriole replication(GO:0007099) |
| 3.4 | 27.3 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 3.4 | 10.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 3.4 | 16.9 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 3.4 | 43.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 3.4 | 10.1 | GO:0045819 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) negative regulation of apoptotic cell clearance(GO:2000426) |
| 3.4 | 40.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 3.3 | 10.0 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 3.3 | 29.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 3.3 | 56.1 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 3.3 | 46.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 3.3 | 6.5 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 3.3 | 13.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 3.3 | 6.5 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 3.3 | 35.8 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 3.2 | 9.7 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 3.2 | 9.6 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of epithelial cell differentiation involved in kidney development(GO:2000698) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 3.2 | 6.3 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 3.2 | 9.5 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 3.1 | 12.6 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 3.1 | 144.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 3.1 | 43.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 3.1 | 9.4 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 3.1 | 15.5 | GO:0072431 | signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 3.1 | 24.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 3.1 | 36.9 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 3.1 | 15.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 3.1 | 24.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 3.1 | 6.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 3.0 | 21.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 3.0 | 15.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 3.0 | 6.0 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 3.0 | 144.1 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 3.0 | 17.9 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 2.9 | 253.0 | GO:0006342 | chromatin silencing(GO:0006342) |
| 2.9 | 11.5 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 2.9 | 5.8 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 2.8 | 8.5 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 2.8 | 5.6 | GO:0061525 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 2.8 | 16.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 2.8 | 16.8 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 2.7 | 5.5 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 2.7 | 13.7 | GO:0090383 | phagosome acidification(GO:0090383) |
| 2.7 | 13.6 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 2.7 | 413.3 | GO:0007059 | chromosome segregation(GO:0007059) |
| 2.6 | 7.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 2.6 | 10.3 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 2.5 | 7.6 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 2.5 | 27.8 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 2.5 | 4.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 2.5 | 7.4 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 2.5 | 14.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 2.4 | 9.8 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 2.4 | 7.3 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 2.4 | 131.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 2.4 | 2.4 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 2.4 | 16.9 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 2.4 | 62.4 | GO:0001709 | cell fate determination(GO:0001709) |
| 2.4 | 2.4 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 2.4 | 66.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 2.4 | 7.1 | GO:0060290 | transdifferentiation(GO:0060290) |
| 2.4 | 4.7 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 2.3 | 7.0 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 2.3 | 97.8 | GO:0048538 | thymus development(GO:0048538) |
| 2.3 | 6.9 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 2.3 | 25.3 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 2.3 | 13.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 2.3 | 9.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 2.3 | 4.5 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 2.3 | 6.8 | GO:0021747 | cochlear nucleus development(GO:0021747) |
| 2.2 | 6.7 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 2.2 | 18.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 2.2 | 15.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 2.2 | 26.6 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 2.2 | 64.1 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 2.2 | 6.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 2.2 | 6.6 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 2.2 | 30.6 | GO:0030320 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) |
| 2.2 | 32.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 2.2 | 8.6 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 2.1 | 2.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 2.1 | 14.9 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 2.1 | 6.3 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 2.1 | 16.8 | GO:0072189 | ureter development(GO:0072189) |
| 2.1 | 2.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 2.1 | 10.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 2.1 | 2.1 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 2.1 | 2.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 2.1 | 10.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 2.1 | 10.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 2.0 | 8.2 | GO:0031297 | replication fork processing(GO:0031297) DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 2.0 | 26.6 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 2.0 | 8.1 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 2.0 | 56.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 2.0 | 4.0 | GO:1903719 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 2.0 | 14.0 | GO:0030035 | microspike assembly(GO:0030035) |
| 2.0 | 35.9 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 2.0 | 8.0 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 2.0 | 21.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 1.9 | 5.8 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 1.9 | 15.0 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 1.9 | 5.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 1.9 | 5.6 | GO:0042637 | catagen(GO:0042637) |
| 1.8 | 31.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 1.8 | 3.7 | GO:0031058 | positive regulation of histone modification(GO:0031058) positive regulation of chromatin modification(GO:1903310) |
| 1.8 | 7.3 | GO:0060325 | head morphogenesis(GO:0060323) face morphogenesis(GO:0060325) |
| 1.8 | 3.7 | GO:0050904 | diapedesis(GO:0050904) |
| 1.8 | 7.3 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 1.8 | 27.1 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 1.8 | 7.2 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 1.8 | 25.0 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 1.8 | 41.0 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 1.8 | 3.6 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 1.8 | 184.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 1.8 | 16.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 1.8 | 15.9 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 1.8 | 3.5 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 1.8 | 8.8 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 1.7 | 3.4 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 1.6 | 9.8 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 1.6 | 3.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 1.6 | 4.7 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 1.6 | 9.3 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 1.5 | 1.5 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 1.5 | 1.5 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 1.5 | 4.5 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 1.5 | 7.4 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 1.5 | 4.4 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 1.5 | 1.5 | GO:2000277 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 1.5 | 29.1 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 1.5 | 4.4 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 1.5 | 1.5 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 1.4 | 1.4 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 1.4 | 4.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 1.4 | 4.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.4 | 8.6 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 1.4 | 1.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 1.4 | 7.1 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 1.4 | 18.4 | GO:0015780 | nucleotide-sugar transport(GO:0015780) |
| 1.4 | 15.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 1.4 | 8.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 1.4 | 15.4 | GO:0070836 | caveola assembly(GO:0070836) |
| 1.4 | 13.9 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 1.4 | 4.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 1.4 | 1.4 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 1.4 | 1.4 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) DNA endoreduplication(GO:0042023) |
| 1.4 | 8.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 1.4 | 5.4 | GO:0015888 | thiamine transport(GO:0015888) |
| 1.4 | 1.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 1.3 | 1.3 | GO:0044805 | late nucleophagy(GO:0044805) |
| 1.3 | 4.0 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 1.3 | 5.4 | GO:0006776 | vitamin A metabolic process(GO:0006776) positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 1.3 | 14.7 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 1.3 | 13.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 1.3 | 6.6 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 1.3 | 11.7 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 1.3 | 3.9 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.3 | 5.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 1.3 | 12.9 | GO:0051451 | myoblast migration(GO:0051451) |
| 1.3 | 3.8 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 1.3 | 5.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 1.3 | 3.8 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 1.3 | 2.5 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 1.3 | 8.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.3 | 11.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 1.3 | 11.3 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 1.2 | 16.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 1.2 | 9.9 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 1.2 | 3.7 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 1.2 | 7.4 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 1.2 | 3.7 | GO:0035672 | transepithelial chloride transport(GO:0030321) oligopeptide transmembrane transport(GO:0035672) |
| 1.2 | 3.7 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 1.2 | 11.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 1.2 | 6.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 1.2 | 3.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 1.2 | 4.8 | GO:0030091 | protein repair(GO:0030091) |
| 1.2 | 3.5 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 1.2 | 15.3 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 1.1 | 6.9 | GO:0060192 | negative regulation of lipase activity(GO:0060192) |
| 1.1 | 52.2 | GO:0007628 | adult walking behavior(GO:0007628) |
| 1.1 | 4.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 1.1 | 33.9 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 1.1 | 9.0 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 1.1 | 2.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 1.1 | 4.5 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) negative regulation of mitochondrial fission(GO:0090258) |
| 1.1 | 23.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 1.1 | 21.9 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of hemostasis(GO:1900048) |
| 1.1 | 2.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 1.1 | 3.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 1.0 | 7.3 | GO:0051195 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 1.0 | 11.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 1.0 | 2.1 | GO:1902022 | L-lysine transport(GO:1902022) |
| 1.0 | 10.2 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 1.0 | 16.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 1.0 | 14.2 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 1.0 | 2.0 | GO:1904152 | regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 1.0 | 11.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 1.0 | 3.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.0 | 6.8 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 1.0 | 2.9 | GO:0070844 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 1.0 | 7.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.9 | 10.3 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.9 | 4.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.9 | 1.9 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.9 | 13.9 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.9 | 9.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.9 | 82.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.9 | 17.5 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.9 | 2.7 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.9 | 3.6 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.9 | 0.9 | GO:0022616 | DNA strand elongation(GO:0022616) |
| 0.9 | 1.8 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.9 | 0.9 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.9 | 7.1 | GO:0090178 | regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.9 | 3.5 | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.9 | 4.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.8 | 5.1 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.8 | 22.2 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.8 | 4.9 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.8 | 3.2 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.8 | 4.0 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.8 | 3.2 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.8 | 6.4 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.8 | 6.3 | GO:1900364 | regulation of mRNA polyadenylation(GO:1900363) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.8 | 7.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.8 | 11.7 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.8 | 3.1 | GO:0002339 | B cell selection(GO:0002339) |
| 0.8 | 4.6 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.8 | 2.3 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.8 | 9.8 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.8 | 1.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.7 | 1.5 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.7 | 6.6 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.7 | 2.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.7 | 5.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.7 | 7.9 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.7 | 2.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.7 | 1.4 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.7 | 2.1 | GO:0072172 | ureteric bud formation(GO:0060676) mesonephric tubule formation(GO:0072172) |
| 0.7 | 7.8 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.7 | 4.2 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.7 | 2.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.7 | 6.9 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.7 | 6.1 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.7 | 6.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.7 | 8.5 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.7 | 3.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.6 | 4.5 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.6 | 3.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.6 | 1.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.6 | 2.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.6 | 13.7 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.6 | 1.2 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.6 | 0.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.6 | 1.8 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.6 | 3.0 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.6 | 6.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.6 | 2.3 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.6 | 5.8 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.6 | 7.6 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.6 | 3.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.6 | 5.8 | GO:0001947 | heart looping(GO:0001947) |
| 0.6 | 12.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.6 | 3.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.6 | 2.8 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.6 | 10.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.6 | 5.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.6 | 3.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.6 | 9.0 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.6 | 2.2 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.6 | 1.1 | GO:0097168 | mesenchymal stem cell maintenance involved in nephron morphogenesis(GO:0072038) mesenchymal stem cell proliferation(GO:0097168) |
| 0.5 | 2.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.5 | 12.6 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.5 | 6.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.5 | 16.7 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.5 | 5.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.5 | 1.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.5 | 23.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.5 | 3.6 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.5 | 5.7 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.5 | 5.2 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.5 | 3.5 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.5 | 1.0 | GO:0009838 | abscission(GO:0009838) |
| 0.5 | 3.5 | GO:0019363 | pyridine nucleotide biosynthetic process(GO:0019363) |
| 0.5 | 3.9 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.5 | 13.8 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.5 | 11.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.5 | 6.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.5 | 0.5 | GO:1902591 | single-organism membrane budding(GO:1902591) |
| 0.5 | 10.6 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.5 | 3.4 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.5 | 5.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.5 | 8.0 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.5 | 0.9 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.5 | 28.8 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.5 | 5.4 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.4 | 0.9 | GO:0019441 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.4 | 9.3 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.4 | 7.0 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.4 | 6.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.4 | 3.9 | GO:0000303 | response to superoxide(GO:0000303) adaptive thermogenesis(GO:1990845) |
| 0.4 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.4 | 2.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.4 | 7.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.4 | 7.9 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.4 | 4.5 | GO:0048536 | spleen development(GO:0048536) |
| 0.4 | 1.2 | GO:0009068 | aspartate family amino acid catabolic process(GO:0009068) |
| 0.4 | 4.1 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.4 | 3.7 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.4 | 8.6 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.4 | 0.8 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.4 | 3.3 | GO:0039703 | viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.4 | 5.3 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.4 | 12.4 | GO:0060425 | lung morphogenesis(GO:0060425) |
| 0.4 | 2.0 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.4 | 8.3 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.4 | 8.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.4 | 3.5 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.4 | 1.5 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.4 | 10.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.4 | 4.6 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.4 | 1.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.4 | 4.6 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.4 | 0.8 | GO:1902302 | regulation of potassium ion export(GO:1902302) |
| 0.4 | 5.2 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.4 | 2.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.4 | 1.1 | GO:0045639 | positive regulation of myeloid cell differentiation(GO:0045639) |
| 0.4 | 1.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.4 | 0.4 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.4 | 5.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.4 | 9.3 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.4 | 6.1 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.4 | 5.7 | GO:0036293 | response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) |
| 0.3 | 2.1 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.3 | 3.8 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.3 | 8.4 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.3 | 14.9 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.3 | 1.7 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.3 | 5.5 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.3 | 1.3 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.3 | 12.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.3 | 10.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.3 | 1.3 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.3 | 10.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.3 | 0.7 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.3 | 5.2 | GO:0044243 | multicellular organism catabolic process(GO:0044243) |
| 0.3 | 2.3 | GO:0032366 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.3 | 19.7 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.3 | 9.3 | GO:0046039 | GTP metabolic process(GO:0046039) |
| 0.3 | 0.3 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) negative regulation of gene silencing(GO:0060969) |
| 0.3 | 1.3 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.3 | 1.9 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.3 | 1.9 | GO:0032196 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) transposition(GO:0032196) |
| 0.3 | 5.4 | GO:0009065 | glutamine family amino acid catabolic process(GO:0009065) |
| 0.3 | 1.6 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.3 | 1.9 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.3 | 1.9 | GO:0060438 | trachea development(GO:0060438) |
| 0.3 | 7.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.3 | 0.9 | GO:0032375 | negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) |
| 0.3 | 1.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.3 | 0.3 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.3 | 2.9 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.3 | 6.7 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.3 | 0.9 | GO:0044068 | modulation by symbiont of host cellular process(GO:0044068) |
| 0.3 | 14.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.3 | 3.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.3 | 3.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.3 | 1.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 4.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.3 | 0.6 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.3 | 1.1 | GO:0061623 | glycolytic process from galactose(GO:0061623) |
| 0.3 | 8.2 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.3 | 5.9 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.3 | 4.7 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.3 | 2.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.3 | 0.8 | GO:0042823 | water-soluble vitamin biosynthetic process(GO:0042364) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.3 | 9.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.3 | 2.8 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.3 | 1.0 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 3.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.2 | 0.5 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 1.0 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.2 | 14.5 | GO:0045137 | development of primary sexual characteristics(GO:0045137) |
| 0.2 | 2.1 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.2 | 4.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.2 | 3.0 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) |
| 0.2 | 5.3 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.2 | 0.7 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.2 | 1.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 0.9 | GO:0035617 | positive regulation of synaptic transmission, GABAergic(GO:0032230) stress granule disassembly(GO:0035617) |
| 0.2 | 2.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 1.1 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.2 | 2.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 0.6 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 1.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.2 | 0.8 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 1.9 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 0.4 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.2 | 2.3 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.2 | 4.6 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 0.4 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.2 | 1.3 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) |
| 0.2 | 4.4 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 1.4 | GO:0031274 | regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 1.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.2 | 5.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.2 | 1.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.2 | 5.6 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.2 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.2 | 2.4 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.2 | 1.7 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 6.2 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.2 | 3.1 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.2 | 0.5 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.1 | 0.3 | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress(GO:0036490) |
| 0.1 | 1.7 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.6 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 4.1 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.1 | 1.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.1 | 3.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.2 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 2.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 6.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 2.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.3 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.1 | 4.5 | GO:0060348 | bone development(GO:0060348) |
| 0.1 | 1.3 | GO:0003170 | heart valve development(GO:0003170) |
| 0.1 | 2.3 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 0.3 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.5 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.1 | 1.0 | GO:0015800 | acidic amino acid transport(GO:0015800) L-glutamate transport(GO:0015813) |
| 0.1 | 0.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 2.0 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.1 | 0.6 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 0.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.8 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.8 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 1.0 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.1 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.1 | 1.2 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 3.2 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.1 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 1.1 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.1 | 0.5 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.1 | 5.0 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 3.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 3.4 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.1 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.1 | 2.2 | GO:0000075 | cell cycle checkpoint(GO:0000075) |
| 0.1 | 0.5 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.7 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 1.8 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.3 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 2.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.2 | GO:0019585 | uronic acid metabolic process(GO:0006063) glucuronate metabolic process(GO:0019585) cellular glucuronidation(GO:0052695) |
| 0.1 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:2000744 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.9 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.4 | GO:0019471 | 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 1.0 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.4 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.3 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.3 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.1 | GO:0035646 | endosome to melanosome transport(GO:0035646) pigment accumulation(GO:0043476) cellular pigment accumulation(GO:0043482) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 | 0.2 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.4 | GO:0051099 | positive regulation of binding(GO:0051099) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 82.6 | 412.9 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 42.8 | 214.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 41.0 | 164.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 34.1 | 136.6 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 27.8 | 194.8 | GO:0005818 | aster(GO:0005818) |
| 27.1 | 135.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 25.2 | 125.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 19.8 | 59.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 19.7 | 196.7 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 19.5 | 58.5 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 16.1 | 80.5 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 15.4 | 108.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 15.2 | 45.7 | GO:0000801 | central element(GO:0000801) |
| 14.7 | 250.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 13.7 | 13.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 13.0 | 39.0 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 9.9 | 29.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 9.9 | 128.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 9.3 | 46.3 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 7.5 | 29.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 7.1 | 178.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 7.0 | 7.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 6.3 | 252.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 6.2 | 436.8 | GO:0000786 | nucleosome(GO:0000786) |
| 6.2 | 31.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 6.2 | 74.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 6.0 | 24.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 5.7 | 22.6 | GO:0043293 | apoptosome(GO:0043293) |
| 5.5 | 87.6 | GO:0010369 | chromocenter(GO:0010369) |
| 5.4 | 37.7 | GO:0001940 | male pronucleus(GO:0001940) |
| 5.3 | 10.6 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 5.0 | 30.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 5.0 | 5.0 | GO:0001739 | sex chromatin(GO:0001739) |
| 4.5 | 13.5 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 4.5 | 22.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 4.4 | 17.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 4.4 | 17.4 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 4.3 | 455.3 | GO:0000793 | condensed chromosome(GO:0000793) |
| 4.2 | 29.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 4.2 | 59.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 4.1 | 258.8 | GO:0005657 | replication fork(GO:0005657) |
| 4.0 | 16.2 | GO:0071942 | XPC complex(GO:0071942) |
| 4.0 | 4.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 3.9 | 30.9 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 3.7 | 26.1 | GO:0045120 | pronucleus(GO:0045120) |
| 3.7 | 55.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 3.7 | 14.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 3.6 | 21.5 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 3.5 | 3.5 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 3.5 | 180.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 3.4 | 20.7 | GO:0097513 | myosin II filament(GO:0097513) |
| 3.4 | 27.2 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 3.3 | 83.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 3.3 | 6.6 | GO:0001939 | female pronucleus(GO:0001939) |
| 3.3 | 236.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 3.2 | 16.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 3.2 | 9.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 3.2 | 9.6 | GO:0071914 | prominosome(GO:0071914) |
| 3.2 | 9.5 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 3.1 | 21.4 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 3.0 | 301.7 | GO:0000776 | kinetochore(GO:0000776) |
| 3.0 | 24.4 | GO:0005883 | neurofilament(GO:0005883) |
| 2.8 | 11.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 2.8 | 41.7 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 2.7 | 8.2 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 2.7 | 13.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 2.6 | 7.9 | GO:0031251 | PAN complex(GO:0031251) |
| 2.6 | 28.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 2.6 | 23.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 2.6 | 33.5 | GO:0042555 | MCM complex(GO:0042555) |
| 2.4 | 16.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 2.4 | 11.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 2.4 | 7.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 2.3 | 14.0 | GO:0044305 | calyx of Held(GO:0044305) |
| 2.3 | 9.3 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 2.3 | 27.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 2.3 | 6.8 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 2.3 | 31.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 2.3 | 31.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 2.2 | 4.5 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 2.1 | 32.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 2.1 | 31.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 2.1 | 47.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 2.0 | 16.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 2.0 | 37.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 1.9 | 5.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.9 | 5.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 1.8 | 38.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 1.8 | 3.6 | GO:0005687 | U4 snRNP(GO:0005687) |
| 1.7 | 85.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 1.7 | 31.0 | GO:0000152 | nuclear ubiquitin ligase complex(GO:0000152) |
| 1.7 | 10.2 | GO:0070187 | telosome(GO:0070187) |
| 1.7 | 30.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 1.6 | 4.9 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 1.6 | 495.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 1.6 | 1.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 1.6 | 128.6 | GO:0016605 | PML body(GO:0016605) |
| 1.6 | 115.3 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 1.6 | 17.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 1.6 | 12.6 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 1.6 | 12.4 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 1.5 | 4.6 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 1.5 | 7.4 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 1.5 | 23.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 1.4 | 18.8 | GO:0045180 | basal cortex(GO:0045180) |
| 1.4 | 5.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 1.4 | 141.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 1.4 | 95.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 1.4 | 6.9 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 1.4 | 10.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 1.4 | 9.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.3 | 4.0 | GO:1990597 | TRAF2-GSTP1 complex(GO:0097057) AIP1-IRE1 complex(GO:1990597) |
| 1.3 | 3.8 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 1.3 | 7.6 | GO:0034448 | EGO complex(GO:0034448) |
| 1.2 | 105.8 | GO:0005814 | centriole(GO:0005814) |
| 1.2 | 24.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 1.2 | 6.0 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 1.2 | 1.2 | GO:0044301 | climbing fiber(GO:0044301) |
| 1.2 | 6.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.2 | 8.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 1.2 | 13.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 1.1 | 3.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 1.1 | 16.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 1.1 | 3.3 | GO:0034657 | GID complex(GO:0034657) |
| 1.1 | 4.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 1.1 | 8.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 1.1 | 6.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 1.1 | 8.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 1.0 | 2.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 1.0 | 6.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 1.0 | 60.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 1.0 | 8.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 1.0 | 11.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 1.0 | 8.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 1.0 | 13.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 1.0 | 16.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 1.0 | 15.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 1.0 | 18.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.9 | 67.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.9 | 9.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.9 | 19.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.9 | 7.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.9 | 241.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.9 | 5.3 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.9 | 59.0 | GO:0016234 | inclusion body(GO:0016234) |
| 0.9 | 199.3 | GO:0005694 | chromosome(GO:0005694) |
| 0.9 | 9.6 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.9 | 24.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.9 | 8.6 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.9 | 3.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.8 | 0.8 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.8 | 9.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.8 | 4.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.8 | 5.7 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.8 | 9.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.8 | 3.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.8 | 2.3 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.8 | 7.6 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.7 | 4.4 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.7 | 6.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.7 | 3.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.7 | 4.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.7 | 2.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.7 | 19.3 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.7 | 80.4 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.7 | 11.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.7 | 5.4 | GO:1990752 | microtubule end(GO:1990752) |
| 0.7 | 2.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.6 | 4.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.6 | 3.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.6 | 4.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.6 | 4.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.6 | 5.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.6 | 7.9 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.6 | 5.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.5 | 3.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 3.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.5 | 22.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.5 | 31.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.5 | 10.9 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.5 | 9.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.5 | 2.5 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.5 | 2.0 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.5 | 3.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.5 | 10.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.5 | 3.0 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.5 | 7.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.5 | 0.5 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.5 | 0.9 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.5 | 12.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.5 | 52.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.5 | 35.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.5 | 0.9 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.4 | 1.3 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.4 | 34.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.4 | 2.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.4 | 3.0 | GO:0031415 | NatA complex(GO:0031415) |
| 0.4 | 5.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.4 | 3.8 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.4 | 1.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 10.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.3 | 2.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 19.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.3 | 3.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 3.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 1.6 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.3 | 2.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.3 | 1.9 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.3 | 2.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 1.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.3 | 1.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.3 | 7.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.3 | 1.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.2 | 1.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 3.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.2 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.2 | 7.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.2 | 3.7 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 62.0 | GO:0005813 | centrosome(GO:0005813) |
| 0.2 | 1.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 5.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.2 | 1.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.2 | 4.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 0.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 3.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.2 | 4.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 871.4 | GO:0005634 | nucleus(GO:0005634) |
| 0.2 | 1.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 2.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 3.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 5.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 3.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 7.0 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 2.7 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 0.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.3 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 1.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.7 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 47.5 | 190.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 45.7 | 182.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 37.7 | 37.7 | GO:0097472 | cyclin-dependent protein kinase activity(GO:0097472) |
| 31.6 | 158.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 30.1 | 90.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 28.0 | 112.0 | GO:0031493 | nucleosomal histone binding(GO:0031493) hemi-methylated DNA-binding(GO:0044729) |
| 27.4 | 82.1 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 24.9 | 149.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 22.7 | 136.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 20.3 | 101.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 19.0 | 56.9 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 14.4 | 114.9 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 12.8 | 115.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 11.7 | 23.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 10.4 | 10.4 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 10.3 | 92.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 9.2 | 36.8 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 8.9 | 436.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 8.3 | 50.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 7.9 | 31.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 7.8 | 179.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 7.2 | 64.8 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 7.0 | 28.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 7.0 | 34.8 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 6.8 | 20.5 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 6.7 | 93.7 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 6.6 | 26.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 6.3 | 44.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 6.3 | 18.8 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 6.2 | 49.5 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 6.0 | 84.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 6.0 | 35.9 | GO:0070728 | leucine binding(GO:0070728) |
| 5.7 | 34.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 5.6 | 22.4 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 5.6 | 16.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 5.4 | 16.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 5.3 | 21.1 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 5.2 | 41.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 5.1 | 20.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 5.1 | 81.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 5.1 | 5.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 5.0 | 352.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 5.0 | 19.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 4.9 | 39.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 4.9 | 19.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 4.7 | 14.0 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 4.7 | 88.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 4.5 | 13.5 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 4.4 | 17.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 4.4 | 17.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 4.3 | 12.9 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 4.1 | 12.4 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 4.1 | 16.3 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 4.0 | 16.1 | GO:0004096 | aminoacylase activity(GO:0004046) catalase activity(GO:0004096) |
| 4.0 | 251.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 3.9 | 15.7 | GO:0046790 | virion binding(GO:0046790) |
| 3.8 | 11.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 3.8 | 15.2 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 3.7 | 14.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 3.6 | 126.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 3.6 | 21.5 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 3.5 | 17.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 3.5 | 10.6 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 3.4 | 10.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 3.4 | 10.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 3.4 | 16.9 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 3.3 | 16.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 3.3 | 16.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 3.2 | 9.7 | GO:0032564 | dATP binding(GO:0032564) |
| 3.2 | 151.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 3.2 | 48.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 3.2 | 19.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 3.2 | 34.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 3.1 | 9.4 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 3.1 | 9.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 3.1 | 368.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 3.1 | 33.7 | GO:0008430 | selenium binding(GO:0008430) |
| 3.0 | 21.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 3.0 | 20.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 2.9 | 32.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 2.9 | 11.7 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 2.9 | 17.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 2.8 | 73.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 2.8 | 31.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 2.8 | 16.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 2.7 | 18.6 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 2.5 | 7.6 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 2.5 | 27.3 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 2.4 | 9.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 2.4 | 9.7 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 2.4 | 31.3 | GO:0031386 | protein tag(GO:0031386) |
| 2.4 | 7.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) sodium:inorganic phosphate symporter activity(GO:0015319) |
| 2.4 | 9.6 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 2.4 | 14.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 2.4 | 9.5 | GO:0036435 | deubiquitinase activator activity(GO:0035800) K48-linked polyubiquitin binding(GO:0036435) |
| 2.4 | 42.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 2.3 | 16.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 2.3 | 9.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 2.3 | 38.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 2.3 | 13.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 2.2 | 6.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 2.2 | 13.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 2.2 | 4.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 2.2 | 21.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 2.1 | 8.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 2.1 | 36.2 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 2.1 | 6.4 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 2.1 | 90.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 2.0 | 150.5 | GO:0001047 | core promoter binding(GO:0001047) |
| 2.0 | 12.0 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 2.0 | 8.0 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 2.0 | 45.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 2.0 | 5.9 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 2.0 | 7.8 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 1.9 | 15.5 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 1.9 | 5.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.8 | 9.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 1.8 | 21.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 1.8 | 316.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 1.8 | 7.3 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 1.8 | 1.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 1.8 | 5.4 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 1.8 | 5.3 | GO:0030984 | kininogen binding(GO:0030984) |
| 1.8 | 24.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 1.7 | 40.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.7 | 12.2 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.7 | 17.1 | GO:0070888 | E-box binding(GO:0070888) |
| 1.7 | 26.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 1.6 | 3.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 1.6 | 4.9 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 1.6 | 1.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.6 | 93.5 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 1.6 | 34.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 1.6 | 158.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 1.6 | 4.7 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 1.5 | 275.8 | GO:0042393 | histone binding(GO:0042393) |
| 1.5 | 16.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 1.5 | 19.7 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 1.5 | 4.5 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 1.5 | 13.3 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 1.5 | 45.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 1.5 | 8.9 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 1.5 | 26.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 1.5 | 4.4 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 1.4 | 7.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 1.4 | 305.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 1.4 | 5.6 | GO:2001070 | starch binding(GO:2001070) |
| 1.4 | 20.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 1.4 | 5.5 | GO:0047134 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) protein-disulfide reductase activity(GO:0047134) |
| 1.4 | 6.9 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 1.4 | 375.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 1.4 | 9.5 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 1.4 | 44.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 1.4 | 4.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 1.3 | 54.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 1.3 | 7.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.3 | 6.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 1.3 | 3.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 1.3 | 5.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 1.3 | 8.9 | GO:0015925 | galactosidase activity(GO:0015925) |
| 1.3 | 5.0 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 1.3 | 3.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 1.2 | 14.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 1.2 | 3.7 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 1.2 | 9.6 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 1.2 | 47.7 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 1.2 | 4.6 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 1.2 | 1.2 | GO:0003681 | bent DNA binding(GO:0003681) |
| 1.2 | 9.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 1.1 | 10.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 1.1 | 7.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 1.1 | 3.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.1 | 17.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 1.1 | 5.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 1.1 | 9.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 1.1 | 5.3 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 1.0 | 10.4 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 1.0 | 9.2 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 1.0 | 6.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 1.0 | 66.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 1.0 | 8.0 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 1.0 | 29.9 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 1.0 | 9.0 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 1.0 | 3.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 1.0 | 3.9 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 1.0 | 19.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 1.0 | 5.8 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 1.0 | 2.9 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.9 | 3.7 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.9 | 5.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.9 | 4.5 | GO:0089720 | caspase binding(GO:0089720) |
| 0.9 | 5.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.9 | 12.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.9 | 5.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.8 | 1.7 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.8 | 16.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.8 | 9.2 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.8 | 5.7 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.8 | 18.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.8 | 26.7 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.8 | 2.3 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.8 | 15.8 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.7 | 2.2 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.7 | 7.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.7 | 4.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.7 | 3.7 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.7 | 5.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.7 | 4.4 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.7 | 17.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.7 | 3.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.7 | 3.5 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.7 | 53.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.7 | 4.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.7 | 30.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.7 | 21.0 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.6 | 20.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.6 | 43.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.6 | 1.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.6 | 22.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.6 | 3.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.6 | 3.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.6 | 4.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.6 | 6.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.6 | 2.4 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.6 | 4.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.6 | 3.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.6 | 3.0 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.6 | 2.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.6 | 5.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.6 | 2.3 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.6 | 2.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.6 | 3.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.6 | 6.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.6 | 29.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.6 | 5.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.5 | 2.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.5 | 1.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.5 | 4.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.5 | 5.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 6.7 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.5 | 2.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.5 | 4.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.5 | 6.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.5 | 1.4 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.5 | 1.9 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.5 | 43.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.5 | 9.2 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.5 | 1.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.5 | 1.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.5 | 0.9 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.5 | 29.3 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.4 | 1.8 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.4 | 1.3 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.4 | 12.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.4 | 6.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.4 | 3.4 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.4 | 4.7 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.4 | 63.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.4 | 5.9 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.4 | 8.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.4 | 7.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.4 | 2.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 101.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.4 | 7.0 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.4 | 2.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.4 | 0.8 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.4 | 2.4 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.4 | 14.4 | GO:0003774 | motor activity(GO:0003774) |
| 0.4 | 2.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 2.7 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.4 | 1.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.4 | 30.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.4 | 45.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.4 | 17.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.4 | 1.1 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.4 | 27.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.3 | 28.9 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.3 | 6.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.3 | 1.3 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.3 | 4.1 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.3 | 2.8 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.3 | 6.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 1.9 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.3 | 1.8 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 6.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 4.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.3 | 3.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.3 | 3.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 41.2 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.3 | 3.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.3 | 6.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 11.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.3 | 0.8 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.3 | 2.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.3 | 1.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 1.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 66.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.2 | 2.6 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 0.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.2 | 4.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.2 | 1.5 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.2 | 5.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 1.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.2 | 0.8 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.2 | 0.6 | GO:0030249 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.2 | 0.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 0.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.2 | 1.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.2 | 2.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.2 | 2.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.2 | 17.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.2 | 0.9 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 2.1 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.2 | 0.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.2 | 5.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.2 | 129.7 | GO:0003677 | DNA binding(GO:0003677) |
| 0.1 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 3.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 3.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 43.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 3.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.4 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 5.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 3.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 36.1 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.1 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 2.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 2.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 0.2 | GO:0016215 | acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.3 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 0.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 6.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 2.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 1.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.2 | GO:0046997 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.1 | 0.6 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.3 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 4.5 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 19.7 | 1243.3 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 12.8 | 179.4 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 9.9 | 364.7 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 5.4 | 236.4 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 5.2 | 301.3 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 4.8 | 133.9 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 4.1 | 134.6 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 3.6 | 149.2 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 3.4 | 98.8 | PID_SMAD2_3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 3.2 | 32.5 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 2.8 | 52.7 | PID_MYC_PATHWAY | C-MYC pathway |
| 2.5 | 181.0 | PID_TELOMERASE_PATHWAY | Regulation of Telomerase |
| 2.4 | 7.1 | SA_TRKA_RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 2.4 | 47.0 | PID_FOXM1_PATHWAY | FOXM1 transcription factor network |
| 2.3 | 114.5 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 2.1 | 10.6 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 2.1 | 92.8 | PID_ERBB2_ERBB3_PATHWAY | ErbB2/ErbB3 signaling events |
| 2.1 | 8.4 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 2.1 | 24.8 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 1.9 | 51.3 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 1.6 | 89.9 | PID_E2F_PATHWAY | E2F transcription factor network |
| 1.6 | 52.8 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 1.5 | 1.5 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 1.3 | 38.9 | PID_WNT_NONCANONICAL_PATHWAY | Noncanonical Wnt signaling pathway |
| 1.2 | 37.2 | ST_P38_MAPK_PATHWAY | p38 MAPK Pathway |
| 1.2 | 3.6 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 1.2 | 17.5 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 1.1 | 48.4 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 1.1 | 9.0 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 1.1 | 60.3 | PID_MET_PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 1.1 | 9.8 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 1.1 | 11.7 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 1.0 | 2.9 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.9 | 11.2 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.9 | 13.9 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.9 | 50.3 | PID_CDC42_PATHWAY | CDC42 signaling events |
| 0.8 | 51.7 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.8 | 18.6 | PID_HEDGEHOG_GLI_PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.8 | 29.3 | PID_THROMBIN_PAR1_PATHWAY | PAR1-mediated thrombin signaling events |
| 0.7 | 6.7 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.7 | 15.4 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.7 | 10.5 | ST_PHOSPHOINOSITIDE_3_KINASE_PATHWAY | PI3K Pathway |
| 0.7 | 8.7 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.6 | 36.5 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.6 | 9.6 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.6 | 6.3 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.6 | 2.8 | PID_IL23_PATHWAY | IL23-mediated signaling events |
| 0.5 | 6.6 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.5 | 11.1 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.5 | 9.2 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.5 | 24.0 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.5 | 17.8 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.4 | 16.9 | PID_HDAC_CLASSI_PATHWAY | Signaling events mediated by HDAC Class I |
| 0.4 | 13.3 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.4 | 12.3 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.4 | 6.8 | PID_HNF3A_PATHWAY | FOXA1 transcription factor network |
| 0.4 | 2.1 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.3 | 8.0 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.3 | 6.2 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.3 | 11.7 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.3 | 1.9 | PID_AVB3_INTEGRIN_PATHWAY | Integrins in angiogenesis |
| 0.3 | 3.5 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.3 | 3.5 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 3.7 | PID_INTEGRIN_CS_PATHWAY | Integrin family cell surface interactions |
| 0.3 | 4.3 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.2 | 6.5 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.2 | 4.2 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.4 | PID_TCR_CALCIUM_PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.6 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 4.5 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.9 | SIG_PIP3_SIGNALING_IN_B_LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 1.6 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.4 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.5 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.7 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.8 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.1 | PID_IL8_CXCR2_PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.1 | 375.1 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 15.8 | 47.4 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 15.6 | 249.1 | REACTOME_ASSOCIATION_OF_LICENSING_FACTORS_WITH_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 11.5 | 11.5 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 11.0 | 296.7 | REACTOME_KINESINS | Genes involved in Kinesins |
| 10.7 | 236.3 | REACTOME_APC_CDC20_MEDIATED_DEGRADATION_OF_NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 10.5 | 167.9 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 9.6 | 86.1 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 8.0 | 770.9 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 7.5 | 90.6 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 7.2 | 374.3 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 6.9 | 83.1 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 6.4 | 89.0 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 5.7 | 113.9 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 5.0 | 49.5 | REACTOME_HYALURONAN_UPTAKE_AND_DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 4.6 | 73.3 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 4.3 | 349.7 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 4.0 | 44.3 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 3.9 | 82.9 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 3.7 | 7.4 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 3.5 | 79.8 | REACTOME_ACTIVATION_OF_THE_PRE_REPLICATIVE_COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 3.1 | 79.4 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 3.0 | 21.3 | REACTOME_ENDOGENOUS_STEROLS | Genes involved in Endogenous sterols |
| 2.3 | 25.3 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 2.3 | 125.9 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 2.2 | 9.0 | REACTOME_G_BETA_GAMMA_SIGNALLING_THROUGH_PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 2.2 | 57.0 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 2.0 | 22.3 | REACTOME_DEPOSITION_OF_NEW_CENPA_CONTAINING_NUCLEOSOMES_AT_THE_CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 1.7 | 22.6 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 1.7 | 89.5 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 1.6 | 16.2 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 1.6 | 140.4 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 1.6 | 8.0 | REACTOME_CELL_DEATH_SIGNALLING_VIA_NRAGE_NRIF_AND_NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 1.6 | 55.1 | REACTOME_GLUCOSE_TRANSPORT | Genes involved in Glucose transport |
| 1.4 | 58.3 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 1.4 | 9.6 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 1.3 | 18.3 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 1.2 | 13.4 | REACTOME_BASE_FREE_SUGAR_PHOSPHATE_REMOVAL_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 1.2 | 33.7 | REACTOME_REGULATION_OF_BETA_CELL_DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 1.2 | 33.3 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 1.1 | 21.7 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.1 | 21.6 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 1.1 | 83.8 | REACTOME_GLUCOSE_METABOLISM | Genes involved in Glucose metabolism |
| 1.1 | 23.3 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 1.1 | 10.5 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 1.0 | 15.6 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 1.0 | 64.3 | REACTOME_NOTCH1_INTRACELLULAR_DOMAIN_REGULATES_TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.9 | 6.3 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.9 | 20.6 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.9 | 7.9 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.8 | 16.1 | REACTOME_REGULATION_OF_MRNA_STABILITY_BY_PROTEINS_THAT_BIND_AU_RICH_ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.8 | 4.2 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.8 | 3.3 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 0.8 | 4.0 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.8 | 8.8 | REACTOME_SIGNALING_BY_EGFR_IN_CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.8 | 11.0 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.8 | 2.4 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.7 | 8.6 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.7 | 10.5 | REACTOME_ADENYLATE_CYCLASE_INHIBITORY_PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.7 | 24.3 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.6 | 10.9 | REACTOME_REGULATION_OF_AMPK_ACTIVITY_VIA_LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.6 | 16.9 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.6 | 16.2 | REACTOME_ANTIGEN_ACTIVATES_B_CELL_RECEPTOR_LEADING_TO_GENERATION_OF_SECOND_MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.6 | 7.0 | REACTOME_N_GLYCAN_TRIMMING_IN_THE_ER_AND_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.6 | 5.1 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.6 | 15.4 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.5 | 7.1 | REACTOME_VITAMIN_B5_PANTOTHENATE_METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.5 | 40.5 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.5 | 19.8 | REACTOME_MRNA_3_END_PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.5 | 4.2 | REACTOME_THROMBIN_SIGNALLING_THROUGH_PROTEINASE_ACTIVATED_RECEPTORS_PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.5 | 3.7 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.4 | 6.7 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.4 | 7.4 | REACTOME_REGULATION_OF_HYPOXIA_INDUCIBLE_FACTOR_HIF_BY_OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.4 | 6.5 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.4 | 19.4 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.4 | 7.5 | REACTOME_PERK_REGULATED_GENE_EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.3 | 2.7 | REACTOME_G2_M_CHECKPOINTS | Genes involved in G2/M Checkpoints |
| 0.3 | 2.4 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.3 | 3.3 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.3 | 2.9 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.3 | 3.7 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_LATE_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.3 | 26.5 | REACTOME_GASTRIN_CREB_SIGNALLING_PATHWAY_VIA_PKC_AND_MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.3 | 10.4 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 6.0 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 0.7 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.2 | 35.8 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.2 | 11.3 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 3.6 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 2.6 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 1.8 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 1.1 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 1.9 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 1.8 | REACTOME_BILE_ACID_AND_BILE_SALT_METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.2 | 0.8 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 3.4 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 3.8 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 9.4 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.2 | REACTOME_POST_TRANSLATIONAL_MODIFICATION_SYNTHESIS_OF_GPI_ANCHORED_PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 3.9 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.9 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.4 | REACTOME_ACTIVATED_TAK1_MEDIATES_P38_MAPK_ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 0.9 | REACTOME_TRYPTOPHAN_CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 4.2 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 13.8 | REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.1 | 2.2 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.1 | 6.4 | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 1.3 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME_NUCLEAR_SIGNALING_BY_ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.1 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 2.1 | REACTOME_TRANS_GOLGI_NETWORK_VESICLE_BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.2 | REACTOME_GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.8 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_2_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.5 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME_PYRIMIDINE_METABOLISM | Genes involved in Pyrimidine metabolism |