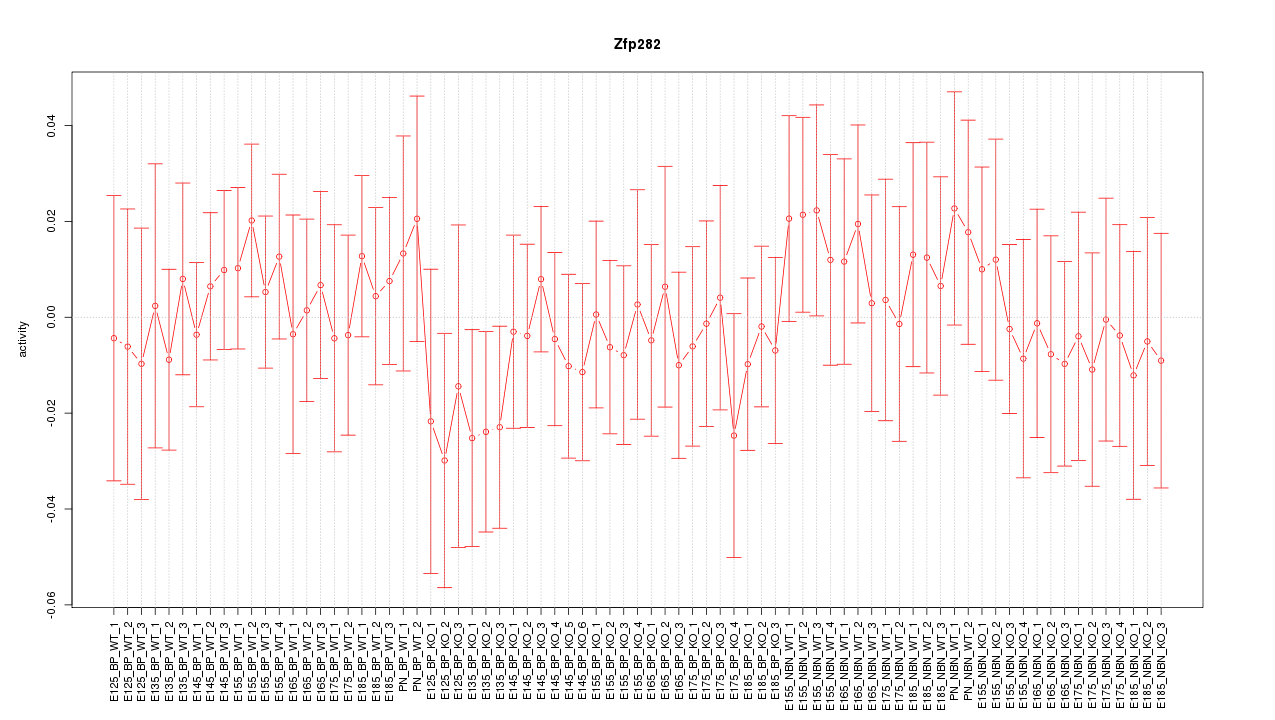
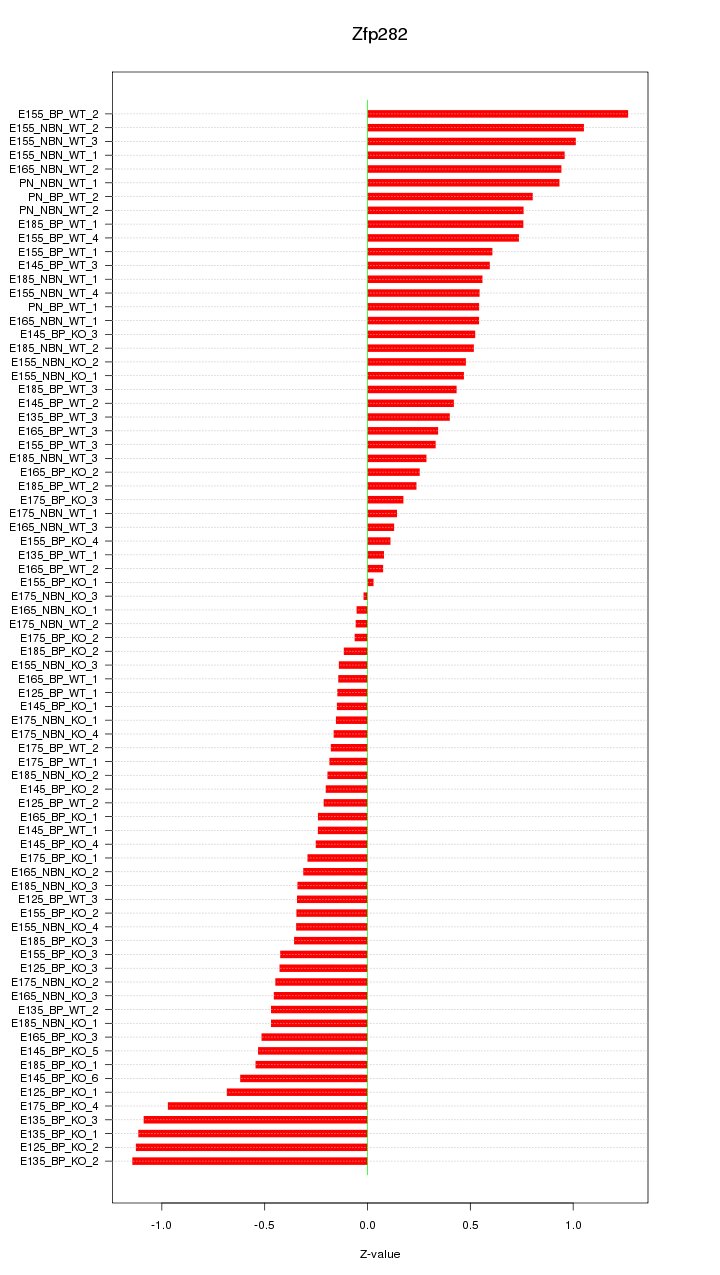
Motif ID: Zfp282
Z-value: 0.545

Transcription factors associated with Zfp282:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp282 | ENSMUSG00000025821.9 | Zfp282 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp282 | mm10_v2_chr6_+_47877204_47877204 | -0.10 | 4.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0042097 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.5 | 2.0 | GO:0009597 | detection of virus(GO:0009597) |
| 0.5 | 2.4 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.4 | 1.3 | GO:0048389 | signal transduction by trans-phosphorylation(GO:0023016) limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) kidney smooth muscle tissue development(GO:0072194) |
| 0.4 | 1.2 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.4 | 1.6 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.3 | 1.0 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.3 | 2.2 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.3 | 1.6 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 3.7 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.2 | 0.9 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.2 | 1.5 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.4 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.7 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.5 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.7 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.4 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.7 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.5 | GO:0086028 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.1 | 3.7 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.2 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.5 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 1.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 1.1 | GO:0042993 | positive regulation of transcription factor import into nucleus(GO:0042993) |
| 0.0 | 1.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 1.8 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 3.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 1.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.5 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 2.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.9 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 2.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.3 | 1.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.3 | 1.6 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.2 | 0.7 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.2 | 1.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.5 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.1 | 0.2 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.9 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 2.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.0 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 3.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.1 | 3.9 | PID_TRAIL_PATHWAY | TRAIL signaling pathway |
| 0.1 | 1.6 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.2 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.2 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 1.8 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | REACTOME_TRAF6_MEDIATED_NFKB_ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 3.5 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.0 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 0.9 | REACTOME_STEROID_HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |