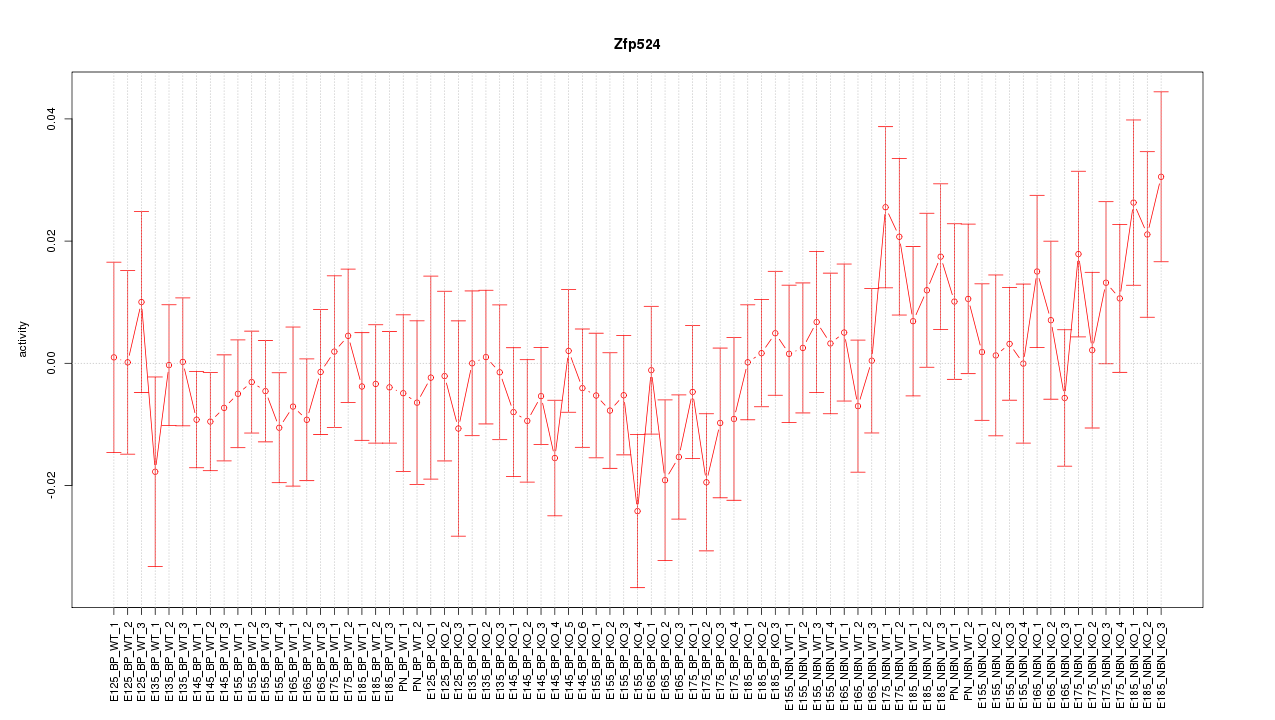
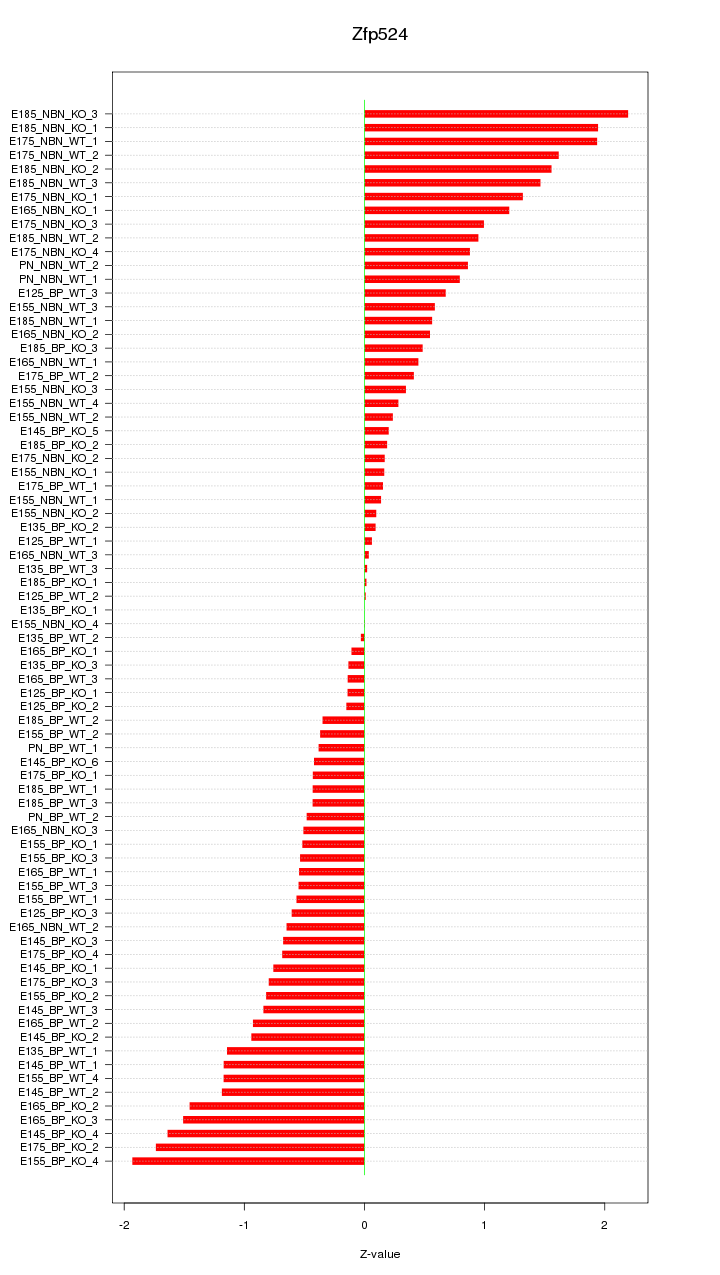
Motif ID: Zfp524
Z-value: 0.866
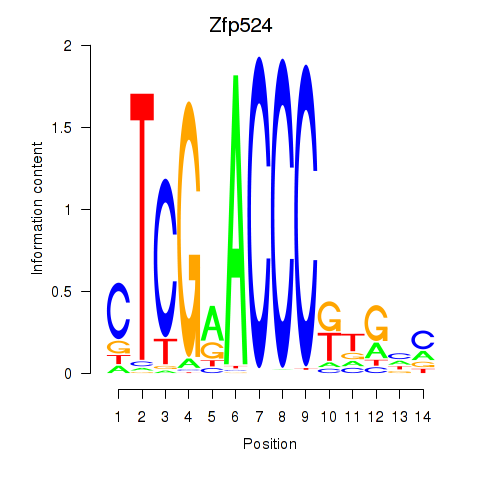
Transcription factors associated with Zfp524:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp524 | ENSMUSG00000051184.6 | Zfp524 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp524 | mm10_v2_chr7_+_5015466_5015509 | -0.55 | 2.3e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 1.9 | 9.3 | GO:2000065 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 1.8 | 11.0 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 1.6 | 8.2 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 1.6 | 4.7 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 1.5 | 5.9 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) thermoception(GO:0050955) |
| 1.3 | 4.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 1.2 | 4.9 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.0 | 3.9 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.9 | 2.8 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.9 | 5.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.8 | 2.5 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.8 | 3.3 | GO:0031622 | positive regulation of fever generation(GO:0031622) |
| 0.8 | 2.5 | GO:0072194 | sensory perception of touch(GO:0050975) kidney smooth muscle tissue development(GO:0072194) |
| 0.8 | 3.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.7 | 2.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.7 | 2.9 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.7 | 2.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.7 | 2.0 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.6 | 3.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.6 | 3.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.6 | 1.9 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.6 | 13.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.6 | 1.9 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.6 | 1.8 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.6 | 4.8 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.6 | 1.8 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.6 | 4.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.6 | 2.3 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.5 | 1.6 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.5 | 6.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.5 | 4.6 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.5 | 13.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.5 | 1.4 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) regulation of microglial cell activation(GO:1903978) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.5 | 10.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.5 | 2.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.4 | 4.4 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.4 | 1.3 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.4 | 1.7 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.4 | 2.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.4 | 0.4 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905024) |
| 0.4 | 1.9 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.4 | 1.1 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.4 | 8.9 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.4 | 2.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.4 | 2.2 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.4 | 1.1 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.4 | 0.7 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.3 | 2.1 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) leading edge cell differentiation(GO:0035026) regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 | 1.6 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.3 | 1.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.3 | 2.3 | GO:1903874 | ferrous iron transmembrane transport(GO:1903874) |
| 0.3 | 2.6 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.3 | 1.6 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.3 | 3.4 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.3 | 2.8 | GO:0043084 | penile erection(GO:0043084) |
| 0.3 | 0.8 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.3 | 2.0 | GO:0032811 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.3 | 1.4 | GO:0031133 | cellular magnesium ion homeostasis(GO:0010961) regulation of axon diameter(GO:0031133) |
| 0.3 | 1.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 1.4 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) |
| 0.3 | 2.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.3 | 2.2 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.3 | 1.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.3 | 1.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.3 | 2.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 2.6 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 | 0.7 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.2 | 2.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 3.8 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.2 | 0.9 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 | 3.3 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.2 | 1.5 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 1.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.2 | 0.6 | GO:2000850 | protein-chromophore linkage(GO:0018298) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.2 | 3.7 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.2 | 0.6 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 2.5 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 0.8 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 1.4 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.2 | 0.8 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.2 | 5.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 3.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 1.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 16.1 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.2 | 1.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.2 | 2.3 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.2 | 1.0 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.2 | 2.7 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.2 | 0.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.2 | 1.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 1.6 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.2 | 1.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 5.0 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.2 | 0.5 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.7 | GO:1905169 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.1 | 1.0 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.9 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.5 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.4 | GO:0032240 | RNA import into nucleus(GO:0006404) mRNA export from nucleus in response to heat stress(GO:0031990) negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.8 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 1.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.8 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 1.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.9 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 1.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 1.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 3.1 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.1 | 0.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.6 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 1.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.4 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 1.1 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.1 | 1.9 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.3 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 0.1 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.5 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 2.3 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.8 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 1.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.3 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.1 | 0.4 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.4 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.2 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 2.7 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 0.3 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.3 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.1 | 0.6 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.6 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 3.1 | GO:0090102 | cochlea development(GO:0090102) |
| 0.1 | 0.2 | GO:0009174 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.1 | 0.8 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.5 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.3 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.6 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.9 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 3.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 1.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.9 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.2 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.1 | 1.5 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 1.5 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.1 | 0.2 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.1 | 0.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 1.1 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 0.5 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 0.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.1 | 3.8 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 1.5 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.2 | GO:0010957 | negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 1.1 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.5 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 1.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 1.1 | GO:0042267 | natural killer cell mediated immunity(GO:0002228) natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.8 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.3 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 1.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.8 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.8 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 2.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.2 | GO:0040031 | snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.0 | 0.0 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.7 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 2.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.5 | GO:0048643 | positive regulation of skeletal muscle tissue development(GO:0048643) |
| 0.0 | 0.5 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.2 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.8 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.7 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 1.8 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.9 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.5 | GO:0052646 | alditol phosphate metabolic process(GO:0052646) |
| 0.0 | 0.2 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.9 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.6 | GO:0035272 | exocrine system development(GO:0035272) |
| 0.0 | 1.4 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.3 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0071281 | cellular response to iron ion(GO:0071281) |
| 0.0 | 0.0 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.3 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.4 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 0.0 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 1.3 | GO:0007411 | axon guidance(GO:0007411) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.6 | 1.9 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.5 | 2.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.5 | 1.6 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.5 | 3.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.5 | 1.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.5 | 1.8 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.4 | 2.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.4 | 3.7 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.4 | 2.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 2.6 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.4 | 1.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.3 | 7.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.3 | 2.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.3 | 2.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 2.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.3 | 0.9 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.3 | 2.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 0.7 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.2 | 1.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 0.7 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 0.7 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.2 | 1.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.2 | 1.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.2 | 10.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 1.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 6.2 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 1.1 | GO:0042025 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.2 | 12.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 1.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) dendritic spine neck(GO:0044326) |
| 0.2 | 2.6 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 0.5 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.2 | 2.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.2 | 2.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 0.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 1.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 14.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.7 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 3.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.1 | 0.5 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 0.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.9 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.8 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 2.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 19.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 4.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.4 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.5 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 0.5 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 3.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.0 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 3.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 3.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.2 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.4 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.1 | 1.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 3.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.2 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.1 | 1.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.6 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 6.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 17.1 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 1.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.8 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 5.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.2 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 3.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 4.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.7 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) cell-substrate junction(GO:0030055) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 1.0 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.2 | GO:0097708 | cytoplasmic vesicle(GO:0031410) intracellular vesicle(GO:0097708) |
| 0.0 | 0.1 | GO:0031982 | vesicle(GO:0031982) |
| 0.0 | 0.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 1.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.1 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 2.0 | 5.9 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) epinephrine binding(GO:0051379) |
| 1.4 | 4.3 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 1.3 | 5.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.8 | 0.8 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.7 | 5.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.7 | 3.7 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.7 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.7 | 6.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.7 | 2.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.6 | 1.9 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.6 | 7.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.5 | 2.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.5 | 1.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.5 | 3.4 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.5 | 3.9 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.5 | 1.4 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.5 | 1.9 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.5 | 1.4 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.4 | 13.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.4 | 2.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.4 | 3.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 2.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 1.6 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.4 | 2.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.4 | 1.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.4 | 2.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.4 | 2.2 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.4 | 1.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 2.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 2.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.3 | 1.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.3 | 1.0 | GO:0019002 | GMP binding(GO:0019002) |
| 0.3 | 1.9 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.3 | 12.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 6.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 1.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.3 | 1.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.3 | 3.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 4.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.2 | 2.5 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.2 | 2.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.9 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 0.7 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.2 | 0.9 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 4.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 4.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 0.6 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.2 | 3.1 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 2.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.2 | 1.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 3.0 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.2 | 2.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.9 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.2 | 0.9 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 1.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.2 | 2.1 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 4.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 0.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 0.5 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.2 | 1.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.5 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 1.5 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.6 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 1.5 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.8 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 2.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.8 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.5 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 6.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 4.6 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 4.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 1.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.3 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.8 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.7 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.3 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.3 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 7.6 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 1.6 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.9 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 3.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 4.1 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.5 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 1.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 0.3 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 2.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.7 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.9 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 18.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.9 | GO:0008238 | exopeptidase activity(GO:0008238) |
| 0.0 | 1.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 3.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 2.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.3 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 11.2 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.9 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 3.0 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.9 | GO:0008144 | drug binding(GO:0008144) |
| 0.0 | 0.3 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 6.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 2.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.5 | GO:0016853 | isomerase activity(GO:0016853) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.6 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 8.8 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.3 | 11.1 | PID_IL8_CXCR1_PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.3 | 6.2 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.3 | 9.5 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.3 | 4.6 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 7.0 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.2 | 0.9 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.2 | 9.2 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.2 | 2.0 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 7.8 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 1.4 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 15.2 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 3.8 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.1 | 4.7 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 3.6 | PID_ATM_PATHWAY | ATM pathway |
| 0.1 | 4.6 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.1 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.8 | PID_GMCSF_PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.7 | SIG_IL4RECEPTOR_IN_B_LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 2.1 | PID_TGFBR_PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.0 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.0 | 5.2 | NABA_ECM_GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.7 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.0 | 2.0 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID_EPO_PATHWAY | EPO signaling pathway |
| 0.0 | 0.8 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.0 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | ST_GA13_PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.2 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.2 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.5 | PID_NFAT_3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 2.8 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.1 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.6 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.8 | 18.9 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.6 | 15.6 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.5 | 5.9 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.4 | 11.5 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.3 | 8.4 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.3 | 6.0 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 7.1 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.3 | 4.5 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 5.0 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 6.4 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.2 | 1.8 | REACTOME_SIGNALING_BY_ACTIVATED_POINT_MUTANTS_OF_FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 5.4 | REACTOME_NETRIN1_SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 2.0 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.3 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.3 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.4 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.1 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 1.8 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.0 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.4 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.7 | REACTOME_DOPAMINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 1.4 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.9 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 3.7 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.0 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.6 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.8 | REACTOME_MRNA_DECAY_BY_3_TO_5_EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.7 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.5 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 2.6 | REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 0.6 | REACTOME_TRAFFICKING_AND_PROCESSING_OF_ENDOSOMAL_TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.1 | REACTOME_GASTRIN_CREB_SIGNALLING_PATHWAY_VIA_PKC_AND_MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.1 | 2.4 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.7 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.7 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.7 | REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.5 | REACTOME_REGULATORY_RNA_PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 1.0 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME_RECEPTOR_LIGAND_BINDING_INITIATES_THE_SECOND_PROTEOLYTIC_CLEAVAGE_OF_NOTCH_RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 3.7 | REACTOME_CLASS_A1_RHODOPSIN_LIKE_RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.7 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.5 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 2.8 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.4 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.9 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME_E2F_ENABLED_INHIBITION_OF_PRE_REPLICATION_COMPLEX_FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.3 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.8 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME_MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.7 | REACTOME_PYRUVATE_METABOLISM_AND_CITRIC_ACID_TCA_CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.2 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.1 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME_CIRCADIAN_CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.8 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.5 | REACTOME_TRANSMISSION_ACROSS_CHEMICAL_SYNAPSES | Genes involved in Transmission across Chemical Synapses |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_GLUCOKINASE_BY_GLUCOKINASE_REGULATORY_PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.7 | REACTOME_TRANSPORT_OF_GLUCOSE_AND_OTHER_SUGARS_BILE_SALTS_AND_ORGANIC_ACIDS_METAL_IONS_AND_AMINE_COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 0.1 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |