Motif ID: Zfp691
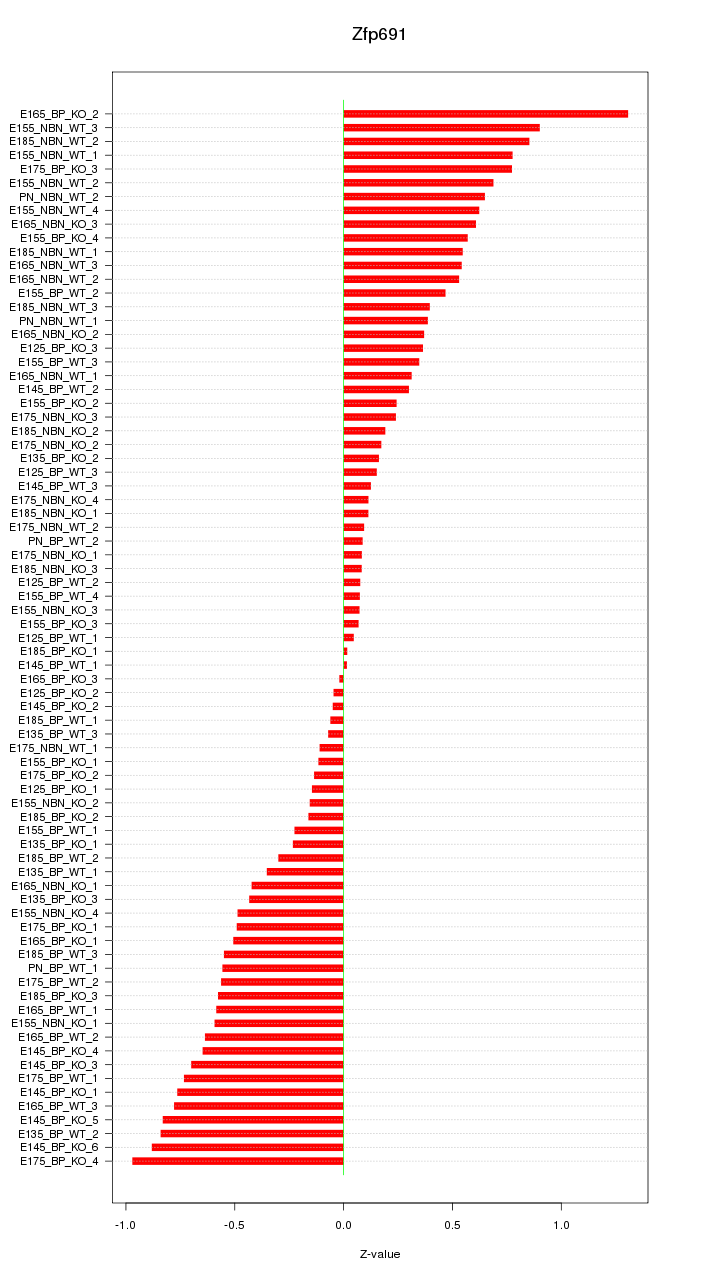
Z-value: 0.489
Transcription factors associated with Zfp691:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp691 | ENSMUSG00000045268.7 | Zfp691 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp691 | mm10_v2_chr4_-_119174178_119174221 | -0.32 | 4.4e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0031038 | myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.8 | 4.9 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.8 | 2.3 | GO:0099548 | trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.5 | 3.2 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.5 | 1.6 | GO:0002489 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) |
| 0.5 | 1.5 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.5 | 2.4 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.3 | 0.8 | GO:1904154 | trimming of terminal mannose on B branch(GO:0036509) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 1.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.2 | 0.7 | GO:1904059 | positive regulation of sensory perception of pain(GO:1904058) regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.6 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.2 | 0.8 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 0.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 1.1 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 1.3 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.1 | 0.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 0.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.6 | GO:0097503 | sialylation(GO:0097503) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.2 | GO:0034312 | diol biosynthetic process(GO:0034312) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.5 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.8 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.7 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 1.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.0 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 2.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.6 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.4 | GO:2000348 | protein linear polyubiquitination(GO:0097039) regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 | 0.7 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 0.3 | GO:1990564 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 1.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.7 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.9 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.5 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.7 | GO:0001881 | receptor recycling(GO:0001881) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 1.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.2 | 0.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 0.7 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 2.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.5 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.7 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 1.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 2.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.8 | GO:0031526 | brush border membrane(GO:0031526) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.4 | 1.6 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.2 | 1.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 1.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 2.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 2.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 1.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.2 | 0.5 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.6 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 2.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.5 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 1.1 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.5 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 2.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 1.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.6 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 1.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.9 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 1.3 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.7 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 4.9 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.5 | REACTOME_IL_7_SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 0.8 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.3 | REACTOME_POST_CHAPERONIN_TUBULIN_FOLDING_PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.6 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.3 | REACTOME_NITRIC_OXIDE_STIMULATES_GUANYLATE_CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 0.6 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.2 | REACTOME_TRNA_AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.9 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.1 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 1.3 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.3 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |