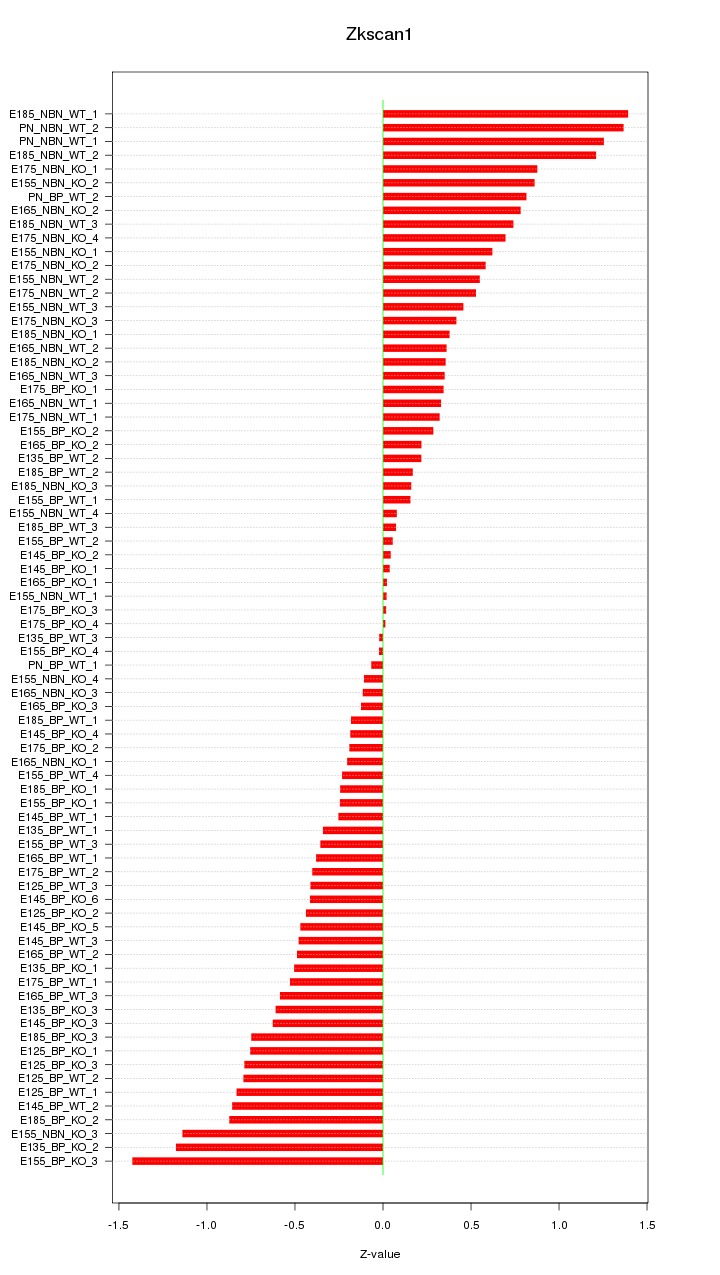
Motif ID: Zkscan1
Z-value: 0.588
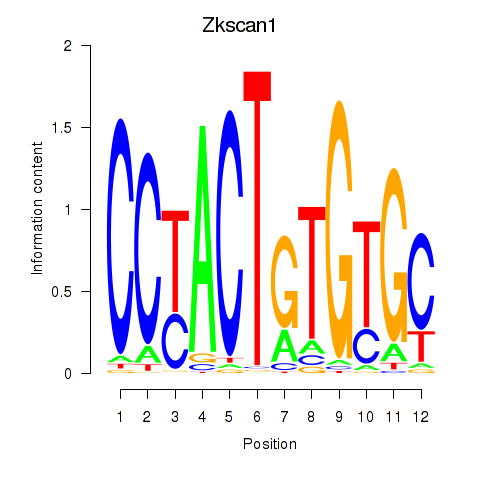
Transcription factors associated with Zkscan1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zkscan1 | ENSMUSG00000029729.6 | Zkscan1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zkscan1 | mm10_v2_chr5_+_138085083_138085147 | -0.23 | 4.7e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.2 | 18.8 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.2 | 4.7 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.1 | 5.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.7 | 2.7 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.6 | 3.2 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.6 | 7.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.6 | 1.8 | GO:2001180 | negative regulation of interleukin-18 production(GO:0032701) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.5 | 1.4 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.4 | 5.9 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.3 | 1.4 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.3 | 4.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.3 | 1.6 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.3 | 3.0 | GO:0046959 | habituation(GO:0046959) |
| 0.3 | 0.9 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.3 | 1.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.2 | 3.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.2 | 0.8 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 0.7 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 | 3.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.9 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.8 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) |
| 0.1 | 0.9 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.1 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.4 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 2.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.8 | GO:1903975 | regulation of glial cell migration(GO:1903975) |
| 0.1 | 3.6 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 0.9 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 1.6 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.1 | 0.2 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.1 | 3.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 2.8 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.1 | 2.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.0 | 1.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.4 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.3 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 6.8 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 4.4 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.0 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.8 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.7 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 1.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 1.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.4 | 1.6 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.4 | 3.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.2 | 2.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.2 | 3.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 17.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.9 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 1.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 6.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.2 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.7 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 4.7 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 2.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.8 | 3.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.6 | 4.7 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.5 | 1.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.3 | 0.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 5.3 | GO:0071949 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) FAD binding(GO:0071949) |
| 0.2 | 1.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 6.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 2.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 7.2 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 2.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.7 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 3.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 3.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 4.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.2 | GO:0046997 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 1.3 | GO:0050811 | GABA-A receptor activity(GO:0004890) GABA receptor binding(GO:0050811) |
| 0.1 | 0.9 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 1.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 3.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 6.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 3.6 | GO:0072509 | divalent inorganic cation transmembrane transporter activity(GO:0072509) |
| 0.0 | 1.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 12.2 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.3 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 5.9 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 3.2 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID_THROMBIN_PAR4_PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.4 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 2.8 | PID_NFAT_3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 2.9 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.1 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.3 | PID_IL8_CXCR2_PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.2 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 2.1 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 3.0 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.0 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.6 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.2 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.4 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.9 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |